Taxa: An Overview of Classification and Major Biological Groupings
Overview and Purpose of Classification
Classification is a way of organizing complex information so that it is easier to process. This normally involves identifying different components that make up the data, then sorting the components into groups based on how similar they are to one another. The degree of similarity is decided based on one or more chosen characteristics. Often groups are arranged into hierarchies to show how different groups and subgroups are related to one another.
In biological classification, groups at different levels in a taxonomic hierarchy are referred to as “ranks.” Ranks that have been commonly used in biology include the following (listed from most general to most specific, with examples given in red):
- Domain Eukarya
- Kingdom Animalia
- Phylum† Chordata
- Class Mammalia
- Order Carnivora
- Family Procyonidae
- Genus Procyon
- Species Procyon lotor
- Genus Procyon
- Family Procyonidae
- Order Carnivora
- Class Mammalia
- Phylum† Chordata
† The term "Division" is usually used instead of "Phylum" when classifying plants and fungi.
.jpg)
Common Raccoon (Procyon lotor)
The basic unit in biological classification is called a taxon (plural: taxa). The word taxon is used to refer to a group of organisms at any level in a taxonomic hierarchy. For example, “animal taxa” refers to all of the groups making up the animals. The genus Procyon is a taxon in the family Procyonidae. Procyon consists of three extant (living) taxa, including Procyon lotor, the Common Raccoon.
The survival of early humans depended on their ability to identify organisms and group them into categories. Even before written language, humans needed to classify organisms into groups based on whether the organisms were edible, poisonous, dangerous, or had other important characteristics (e.g., useful for medical purposes, or for building shelters, making tools, etc.). While initially this information was stored solely within people's brains and was transmitted vocally and/or through gestures, today we have written language and rapidly developing technologies, allowing us to efficiently collect, store and disseminate information. The need for a useful, efficient classification system is becoming ever more important, as the information we accumulate about the biological world continues to grow.
Modern classification systems group organisms based on inferred evolutionary relationships. Taxonomic groups are determined by examining genetic, morphological, and/or anatomical similarities among organisms. The classification systems we use today have several important purposes, including:
- They define and name species. So, in general, different people, in different parts of the world, will refer to the same organism using the same name. This standardization is necessary — otherwise it would be impossible to compare data from different scientific studies. There are of course exceptions — taxonomists do not always agree on how to classify certain organisms.
- Classification systems provide a snapshot of the history of life on earth (albeit a very blurry one with parts missing, gross simplifications, and various errors— the more ancient the time period, the greater the errors and uncertainty). Modern classification systems attempt to show how all organisms that have ever lived on earth are evolutionarily related to one another — a hugely ambitious task.
- Placing organisms into groups that are arranged in a logical hierarchical manner makes it much easier and faster to browse through lists of taxa and find specific information.
- Grouping organisms together that are evolutionarily/biochemically similar to one another is useful for research, technology, and medical purposes. For example, if there is information available for only one taxon in a group — say it’s a rare plant that produces a substance important to science — then you may have a good chance of finding other taxa to study (hopefully ones that aren’t rare) from the same group that produce the same substance. Or say you got bitten by a poisonous spider, which is not well studied, but is known to be closely related to a species for which antivenom is available. Chances are you would benefit from taking the antivenom for the closely related species.
- Classification systems are necessary in order to document, study, and conserve earth’s biodiversity. A species must be described and named first, before it can be recognized as rare and protected from extinction.
- Kingdom Animalia
How Organisms are Classified
The earliest known classification systems placed organisms into groups based on their utility to humans. However, by the 17th century, it became increasingly more common for people to classify life forms using “natural classification”; this involved placing organisms into groups based on shared morphological or anatomical traits, so as to reflect the “order of nature” (Rouhan and Gaudeul 2014). Some advances in taxonomy that followed included the adoption of the binomial scientific name (a two-part Latin name used to refer to a species), as used and popularized by the Swedish taxonomist Carl Linnaeus; and the usage of various hierarchical ranks (e.g., family, genus, species, etc.). At each level in a taxonomic hierarchy, a different characteristic or set of characteristics was used to separate taxa into groups, with all taxa within a given group sharing the same phenotype (appearance) with respect to the chosen characteristic. An attempt was made to classify organisms based on their “natural order” as opposed to using arbitrary characteristics (artificial classification) by applying a principle known as “subordination of characters.” Under the subordination of characters, the traits considered to be most essential to the function of an organism were used to group taxa at the highest level of a taxonomic hierarchy, and increasingly more subordinate traits were used to classify organisms into groups when moving down the taxonomic hierarchy.
A major change in biological classification occurred after 1859, when Charles Darwin published “On the Origin of Species,” a revolutionary book that played a major role in the popularization and acceptance of the theory of evolution. Previously, taxonomists attempted to classify organisms based on the "order of nature" — essentially a biological order as determined by God. It was commonly believed that all organisms came into being at the same time through a creation event — the idea of evolution had not yet been well developed or accepted. However, following the publication of Darwin's book, the theory of evolution received increasingly wider acceptance and support, and scientists began utilizing classification systems based on inferred evolutionary relationships.
Today scientists use a variety of methods and models to determine probable evolutionary relationships. These hypothesized relationships are shown in "phylogenetic trees." Often trees are reconstructed based on comparisons of molecular data (e.g. DNA, RNA, or protein sequences) for the species being evaluated. Although the outcome of these trees may vary depending on the methods and data used to reconstruct them, phylogenetic trees follow a common format:

A simple phylogenetic tree or subtree, drawn in two slightly different ways. This tree shows hypothesized evolutionary relationships between Taxa A, B, C, and D. The internal nodes in this diagram (illustrated here using colored dots) represent ancestral species that gave rise to divergent child species. Taxa that share a common most recent ancestor (i.e., are connected via a single internal node) are considered to be "sister taxa" or "sister groups." For example, in this tree, Taxa A and Taxa B are sister taxa. Monophyletic groups, or clades, consist of all taxa descended from a common ancestor, as well as that ancestor (for example, one clade shown consists of Taxa A and B and the ancestor represented by the green dot). This phylogenetic tree contains two clades that are nested within a larger clade (i.e., the clade consisting of Taxa A and B and their most recent common ancestor, and the clade consisting of Taxa C and D and their most recent common ancestor are nested in a clade that consists of both of those clades plus their most recent common ancestor [represented by the blue dot]).

A simple phylogenetic tree or subtree, drawn in two slightly different ways. This tree shows hypothesized evolutionary relationships between Taxa A, B, C, and D. The internal nodes in this diagram (illustrated here using colored dots) represent ancestral species that gave rise to divergent child species. Taxa that share a common most recent ancestor (i.e., are connected via a single internal node) are considered to be "sister taxa" or "sister groups." For example, in this tree, Taxa A and Taxa B are sister taxa. Monophyletic groups, or clades, consist of all taxa descended from a common ancestor, as well as that ancestor (for example, one clade shown consists of Taxa A and B and the ancestor represented by the green dot). This phylogenetic tree contains two clades that are nested within a larger clade (i.e., the clade consisting of Taxa A and B and their most recent common ancestor, and the clade consisting of Taxa C and D and their most recent common ancestor are nested in a clade that consists of both of those clades plus their most recent common ancestor [represented by the blue dot]).

A simple phylogenetic tree or subtree, drawn in two slightly different ways. This tree shows hypothesized evolutionary relationships between Taxa A, B, C, and D. The internal nodes in this diagram (illustrated here using colored dots) represent ancestral species that gave rise to divergent child species. Taxa that share a common most recent ancestor (i.e., are connected via a single internal node) are considered to be "sister taxa" or "sister groups." For example, in this tree, Taxa A and Taxa B are sister taxa. Monophyletic groups, or clades, consist of all taxa descended from a common ancestor, as well as that ancestor (for example, one clade shown consists of Taxa A and B and the ancestor represented by the green dot). This phylogenetic tree contains two clades that are nested within a larger clade (i.e., the clade consisting of Taxa A and B and their most recent common ancestor, and the clade consisting of Taxa C and D and their most recent common ancestor are nested in a clade that consists of both of those clades plus their most recent common ancestor [represented by the blue dot]).
The most recent taxa (i.e., present day taxa) are shown at the ends of the branches on the tree (i.e., at the external nodes). Each branching point (known as an internal node) represents a hypothetical extinct ancestor, shared by the taxa that diverge from that node. The divergences (bifurcations) shown in these diagrams represent speciation events. In some phylogenetic trees, the length of the branch corresponds to the amount of divergence (genetic change) that has occurred. Phylogenetic trees show the direction of time— as you move from the tips of the branches towards the base or root of the tree, increasingly more ancient taxa are represented at the internal nodes.
From phylogenetic trees, one can identify groupings known as clades. A clade is a taxon (group) that is considered to be “monophyletic” — its members all share a common ancestor, and that ancestor is not shared with any other group. A clade includes the most recent common ancestor for the group and all of that ancestor's descendants. Like taxonomic ranks, clades can be hierarchical — they can be identified at a number of different levels in a phylogenetic tree.
Status of Biological Classification
Biological classification is currently in a state of flux. More and more, scientists are using molecular data, rather than phenotypic traits, to classify organisms. New molecular methods are continually being developed to determine the phylogenetic and taxonomic placement of living things. Increasingly, there is a movement toward the delineation of taxa that are monophyletic (i.e., consist of clades). However, current codes of nomenclature are rank-based (i.e., taxa are placed into hierarchical ranks of kingdom, phylum, class, etc.). The problem with biological ranks is that they do not necessarily consist of clades, and many clades don’t match up with taxonomic ranks— there isn't a one-to-one correspondence between them. To address a lack of guidance for the naming of clades, a draft International Code of Phylogenetic Nomenclature (PhyloCode) was created in 1998 (last updated in 2010) (CPN 2010). Under the PhyloCode, taxa are named in a manner that reflects their phylogeny. The PhyloCode does not deal with ranks. The categories of “species” and “clade” are not considered to be ranks but rather “different kinds of biological entities.” As stated in the preface to the PhyloCode: “…the PhyloCode reflects a philosophical shift from naming species and subsequently classifying them (i.e., into higher taxa) to naming both species and clades.”
Some Problems with Phylogenetic Classification
Phylogenetics is a relatively new science that is still being actively developed. Evolution is an incredibly complex process and attempting to "reconstruct" this process is not an easy task, especially given limited information. The inferred evolutionary relationships shown in phylogenetic trees may vary depending on the genetic datasets that the trees are based upon and the methods and models used. Phylogenetic analyses are very complicated and there are many opportunities for errors or biases to occur, which could lead to incorrect results. Phylogenetic models rely on simplifying assumptions and those assumptions may not apply to the data being evaluated. Phylogenetic studies are especially prone to error when they attempt to determine "deep phylogenies," i.e., evolutionary relationships between very distantly related groups near the root of the tree of life (e.g., evolutionary relationships between Bacteria, Archaea and Eukarya). Because of the very long time that has passed since the divergence of such groups, these analyses involve the difficult task of teasing out a highly attenuated evolutionary signal from a lot of noise, and thus they are much more subject to artifacts (Gouy et al. 2015). Over time, as scientists continue to improve phylogenetic methods and models, they should be able to achieve more consistent and accurate phylogenies, although our reconstructions of evolutionary events will always be associated with a certain level of uncertainty. Some reconstructions, including those of deep phylogenies, may remain highly uncertain.
Tree vs. Net-like Patterns of Evolution
Evolution is typically modeled using a bifurcating "tree" pattern. This pattern assumes that genetic material is passed from parent to offspring, and thus when speciation occurs, from parent to child species. There is therefore a vertical transfer of genes (i.e., when looking at a tree diagram— assuming it's vertically oriented—genes are transferred from an ancestor to its direct descendent in a vertical direction). This assumption, that genes are transferred vertically in a tree-like pattern, is largely true. However, in some cases genes can be transferred or inherited in a manner that leads to a net-like or "reticulate" pattern of branching. For example, in certain instances genes may be transferred horizontally, i.e., from one individual to another individual of the same or different species, where the individual receiving the genes is not the offspring of the one donating the genes.
Horizontal gene transfer can occur in a number of ways. For example, a virus may insert itself into the DNA of a host organism, and in the process leave behind viral genes that become incorporated into the host’s genome. If the infected cell is a germ cell (or otherwise belongs to a line of cells that will get passed to descendants), the viral genes will become a permanent part of the host species’ genome. Bacteria have various methods of acquiring genes, including, under some circumstances, the ability to uptake DNA that they happen to encounter in their environment. Horizontal gene transfer may also occur as the result of an endosymbiosis (a situation where one organism lives inside another). For example, one type of organism (say a cyanobacterium) may become engulfed by another type of organism (say an amoeba), with the engulfed organism surviving inside the other cell. If this relationship persists—because it is beneficial at least to the host organism—then the genomes of the two organisms may merge, at least in part, creating a new type of organism.
Hybridization (the interbreeding between individuals of different species) also results in net-like patterns of evolution. Some species arise as a result of hybridization events (e.g., Mavárez et al. 2006), and consequently have genes that originated from two lines of descent. Also genes from one lineage can get transferred to another if hybrid individuals mate with individuals from one of the parent species (in a process known as introgression).
Net-like patterns of gene transfer can greatly complicate the drawing of phylogenies and the classification of living things. Horizontal gene transfer, which is common in bacteria and other microorganisms, can confound attempts at identifying direct lines of descent, for example if some genes in an organism’s genome originated from bacteria, other genes were acquired from archaea or fungi, etc. Symbiotic life forms, which consist of two or more different organisms living together in close association, also present challenges to phylogenetics and classification. In the case of lichens, which are a symbiosis of algae (or cyanobacteria) and fungi, it is not clear how they should be classified — under algae or fungi? Conventionally, lichens are classified based on the taxonomy of the fungal partner in the symbiosis. However, more recently, it has been discovered that some lichens contain more than one unrelated fungal partner— from different divisions (Spribille et al. 2016)— making classification all the more challenging.
Currently, most phylogenetic analyses ignore net-like branching phenomena and diagram phylogenies using trees. However, given the prevalence of horizontal gene transfer in certain taxa, some scientists believe that the tree of life model should be replaced with a more expansive “network” of life model (Bapteste et al. 2009, 2013; Doolittle 1999; Kunin et al. 2005). According to Philippe et al. (2011), "...phylogenetic trees [represent] only pragmatic approximations which will probably be replaced by phylogenetic networks in the long term (particularly for unicellular organisms)."
Major Biological Groupings
This website groups organisms based on the three cellular domains first described by Carl Woese and colleagues: Bacteria, Archaea and Eukarya (Woese and Fox 1977, Woese et al. 1990). In addition to the three cellular domains, a fourth category is shown on this website: Acellular Parasites. Acellular parasites form part of the gene pool of life on earth. They parasitize and may exchange genetic material with cellular life forms, and thus are important drivers in the evolution of life.
The four basic biological groupings — Acellular Parasites, Bacteria, Archaea, and Eukarya — are described below. Information is briefly summarized for the Acellular Parasites, Bacteria, and Archaea. More details about these groups can be found on their respective web pages. The Eukarya, which are the prime focus of this website, and which are not covered as a group on a separate web page, are discussed in depth. The description of eukaryotes includes extensive information about the other cellular life forms, as they are all inextricably linked.
Acellular Parasites
Acellular parasites are obligate, intracellular parasites that generally consist solely of genetic material (DNA or RNA) or a combination of genetic material and proteins. Examples include viruses and transposons. The acellular parasites are believed to have many different evolutionary origins. This is evident in part by the fact that they don’t all possess the same type of genetic material. The best studied group of acellular parasites, the viruses, do not have a single gene that is shared by all members of the group. In contrast, all cellular life (i.e., spanning the three domains) share core genes, which were likely present in the Last Universal Common Ancestor (LUCA). These include genes that code for ribosomal RNA and genes involved in the transfer of genetic information (Harris et al. 2003).
Because different viruses (and acellular parasites in general), lack a common core of genes, it is essentially impossible to trace their probable evolutionary history back to distant ancestors. Genetic data may be useful in determining evolutionary relationships within particular groups of acellular parasites, as closely related organisms do have similar genomes, with many shared nucleotide sequences. However, it is generally impossible to include acellular parasites on a phylogenetic tree of life due to the general lack of homologous genes among different types of acellular parasites, and the lack of commonality between acellular parasites and cellular life forms.
Bacteria
Bacteria are single-celled microscopic organisms. They are ubiquitous in our environment and are among the most common and abundant life forms on earth. As is true of all cellular life, bacteria have a cell membrane, which controls the movement of substances into and out of the cell; ribosomes, which synthesize proteins; and genetic material consisting of double-stranded DNA. In addition, like all other cellular life forms, they extract energy from food and store it in molecules of adenosine triphosphate (ATP). ATP is a ready-to-use source of energy that can be easily transported to wherever it is needed in a cell.
Most bacteria reproduce via a process known as binary fission. In binary fission, a cell divides in half producing two cells. Prior to cell division, the DNA is copied and is segregated so that one copy of the genetic material goes into each new cell. Although for a long time, bacteria have been described as lacking a cytoskeleton (i.e., a network of protein filaments that extend throughout the cytoplasm), this has been found to be false. Bacteria do, in fact, possess homologs of all of the cytoskeletal elements found in the Eukarya (Celler et al. 2013). For example, the protein FtsZ, which is important in bacterial cell division, is a homolog of the cytoskeletal protein tubulin, found in the Eukarya (Makarova et al. 2010). Cytoskeletal proteins have important functions in cell division, cell motility, and other processes.
The genetic material in bacteria is coiled up into a chromosome. Other DNA molecules, in addition to the main chromosome, may be present in a cell (Egan et al. 2005). Some bacteria contain one copy of their chromosome (i.e., they are monoploid), while others may have multiple copies (i.e., are polyploid) (Pecoraro et al. 2011), and the number of copies of the chromosome(s) can vary widely depending on the phase and rate of growth (du Lac et al. 2017, Maldonado et al. 1994). In most cases the chromosomes are circular. The area they occupy within the cell is called the nucleoid. The nucleoid is mostly separate from the cytoplasm, but it is not surrounded by a membrane. Bacteria and archaea are sometimes referred to as “prokaryotes,” or as having "prokaryotic cells," because their DNA is not contained within a membrane-bound compartment (i.e., they lack a nucleus). The prefix “pro” means before and “karyote” is derived from the Greek word káryon, meaning "nut" or “kernel,” in reference to the nucleus. Essentially, prokaryotes were believed to be simpler cells that preceded cells with nuclei. The usage of the term “prokaryote” has always been somewhat controversial, because it identifies a type of organism based on features that the organism lacks (e.g., no nucleus, no membrane-bound organelles, no mitotic division) (Sapp 2005, Woese and Fox 1977). Some also consider usage of the term prokaryote inappropriate given that it is now known that this group is not monophyletic — the bacteria and the archaea are considered to be two very different cellular life forms. On a further note, the lack of a nucleus or nucleus-like structure may not be a defining feature of all prokaryotes, as indicated by some recent studies (e.g., Sagulenko et al. 2017, Lindsay et al. 2001).
Archaea
Like bacteria, the archaea (singular: archaeon) are single-celled microscopic organisms. They are typically associated with extreme environments or anaerobic conditions (i.e., areas with very high temperature and/or acidity, high salinity, or no dissolved oxygen), as they have been isolated from laboratory cultures of samples collected from such environments. However, more recently, using modern molecular biology techniques that don't require culturing— just the analysis of DNA from environmental samples— archaea have been identified from a wide variety of habitats, including ocean waters, lake waters, and soils (Chaban et al. 2006). Archaea are even known to occur in and on the human body (Nkamga et al. 2017, Probst et al. 2013).
When viewed under a microscope the archaea look a lot like bacteria: their main genetic material, occupying an area called the nucleoid, has no membrane separating it from the rest of the cytoplasm in the cell. Also, their cell sizes and shapes resemble those of bacteria. Compared to cells from Domain Eukarya, cells of archaea and bacteria appear simpler, they lack membrane-bound organelles. Like bacteria, archaea reproduce asexually, primarily by binary fission.[1]
Because archaea superficially resemble bacteria, they weren't recognized as unique life forms — distinct from bacteria — until fairly recently. Archaea were discovered to differ dramatically from bacteria in the 1970s, when microbiologist Carl Woese began analyzing the nucleotide sequences of 16S ribosomal RNA in a variety of organisms, including eukaryotes (i.e., Domain Eukarya), bacteria, and methanogens (anaerobic organisms that were previously believed to be a type of bacteria).[2] Based on ribosomal RNA sequence comparisons, Woese and colleagues found that the methanogens (a type of archaea), and later other types of archaea, are apparently no more related to bacteria than they are to eukaryotes; in fact, they are actually more closely related to eukaryotes (Woese and Fox 1977, Woese et al. 1990). The archaea differ from bacteria in a number of other ways, most notably their cell membranes, cell walls, and method of DNA replication (Sojo et al. 2014). Because of these differences, the antibiotics that we routinely use to fight bacterial infection won't work on archaea (Khelaifia and Drancourt 2012). Fortunately, no archaea are known to be pathogenic (Aminov 2013).
Eukarya
Organisms from the Domain Eukarya are commonly referred to as "eukaryotes" or as having "eukaryotic cells". The name is in reference to one of the distinguishing traits of this group — the presence of a true (membrane-bound) nucleus. (The Greek prefix eu- means "good"; in science it is used in the sense of "true" or "genuine"—in this case in respect to a "true" or "genuine" nucleus.) Eukaryotic cells are also characterized by having other membrane-bound organelles, most notably mitochondria (which are present in one form or another in virtually all eukaryotes) and chloroplasts or other plastids (which are present in all photosynthetic eukaryotes). Cells are further compartmentalized by an extensive internal membrane system (called an endomembrane system), which includes the nuclear membrane, the endoplasmic reticulum, the Golgi apparatus, and other internal membrane features.
Eukaryotes have a cytoskeleton, which is composed of actin filaments, microtubules, intermediate filaments, and other, less described, elements (Moseley 2013). Although bacteria and archaea have homologues to the cytoskeletal proteins found in eukaryotes, the eukaryotic cytoskeleton is more complex and dynamic. For example, only eukaryotic cells are known to be capable of phagocytosis (engulfment of large food particles), a complicated actin-dependent process (Yutin et al. 2009).[3] Unlike bacteria and archaea, eukaryotes have motor proteins (Jékely 2014, Wickstead and Gull 2011)—tiny molecular machines powered by ATP that "walk" along cytoskeletal filament tracks. Some motor proteins carry cargo from one part of the cell to another along the cytoskeletal "highway." Others generate force within the cell, causing changes in cell shape (as in phagocytosis), driving eukaryotic cell division, or powering other cellular movement.
Most eukaryotic cells divide (reproduce asexually) through a process known as mitosis, which is unique to eukaryotes. Prior to mitotic cell division, DNA is replicated (during what is known as the S phase of the cell cycle). This is followed by a gap phase in the cycle. Then, at the beginning of the Mitosis phase, the DNA is condensed into visible chromosomes, and a microtubule-based molecular machine known as a spindle appears. The spindle, powered by motor proteins, lines up duplicated chromosomes in the center of the cell and then simultaneously pulls each of the duplicates apart, with one copy going to each end of the cell, thus ensuring an accurate distribution of chromosomes into each daughter cell. While the chromosomes are still migrating to opposite poles of the cell, the cell begins splitting into two, so the segregation of the DNA and the actual cell division (cytokinesis) are closely coupled. In contrast, in the typical bacterium, the replication of DNA and its segregation into daughter nucleoids occurs concurrently, and there is no close link between DNA segregation and cytokinesis (Kuzminov 2013). Some archaea have been found to have a cell cycle similar to that of eukaryotes, with DNA replication and segregation temporally separated by a gap phase, and with DNA segregation and cytokinesis occurring in rapid succession and appearing to be closely linked (Barillà 2016).
In addition to asexual reproduction via mitosis, eukaryotes also reproduce sexually. Sex (the transfer or exchange of genetic material) and reproduction (the creation of offspring) occur in bacteria and archaea, but in those taxa the two processes are entirely separate and unrelated. In eukaryotes the two processes are coupled. Sexual reproduction in eukaryotes involves two main components: (1) a type of cell division known as meiosis, which results in the production of cells called gametes (e.g. sperm and egg) that have half the number of chromosomes as a typical cell; and (2) the fusion of two gametes (and the fusion of their nuclei) to form an offspring cell, which contains the typical number of chromosomes. Most eukaryotes are diploid (have two sets of chromosomes) and their gametes are haploid/monoploid (have one set of chromosomes). Evidence for sexual reproduction has been found for virtually all eukaryotes studied; in addition, there is a growing consensus among scientists that sexual reproduction was present in the Last Eukaryotic Common Ancestor (LECA) (Speijer et al. 2015).
Endosymbiosis Theory

Rhizosolenia setigera: a type of diatom—a single-celled alga from Domain Eukarya. This cell, which has spine-like projections on either end, is filled with organelles called chloroplasts (visible in the photo). In this organism the "chloroplasts" are technically (or more correctly) referred to as complex plastids (or secondary plastids)—they arose as the result of two endosymbiotic events. In the first event, a heterotrophic eukaryotic cell engulfed a cyanobacterium. The cyanobacterium evolved into a chloroplast and the host cell thus acquired the ability to photosynthesize, becoming an alga. In the second event, another heterotrophic eukaryotic cell (an ancient ancestor to the Rhizosolenia) engulfed one of descendents of the alga (which over time diversified to become red algae, green algae and glaucophytes)—in this case a red alga. The red alga became trapped inside the cell, giving rise to the complex plastids visible in this photo.

Rhizosolenia setigera: a type of diatom—a single-celled alga from Domain Eukarya. This cell, which has spine-like projections on either end, is filled with organelles called chloroplasts (visible in the photo). In this organism the "chloroplasts" are technically (or more correctly) referred to as complex plastids (or secondary plastids)—they arose as the result of two endosymbiotic events. In the first event, a heterotrophic eukaryotic cell engulfed a cyanobacterium. The cyanobacterium evolved into a chloroplast and the host cell thus acquired the ability to photosynthesize, becoming an alga. In the second event, another heterotrophic eukaryotic cell (an ancient ancestor to the Rhizosolenia) engulfed one of descendents of the alga (which over time diversified to become red algae, green algae and glaucophytes)—in this case a red alga. The red alga became trapped inside the cell, giving rise to the complex plastids visible in this photo.

Rhizosolenia setigera: a type of diatom—a single-celled alga from Domain Eukarya. This cell, which has spine-like projections on either end, is filled with organelles called chloroplasts (visible in the photo). In this organism the "chloroplasts" are technically (or more correctly) referred to as complex plastids (or secondary plastids)—they arose as the result of two endosymbiotic events. In the first event, a heterotrophic eukaryotic cell engulfed a cyanobacterium. The cyanobacterium evolved into a chloroplast and the host cell thus acquired the ability to photosynthesize, becoming an alga. In the second event, another heterotrophic eukaryotic cell (an ancient ancestor to the Rhizosolenia) engulfed one of descendents of the alga (which over time diversified to become red algae, green algae and glaucophytes)—in this case a red alga. The red alga became trapped inside the cell, giving rise to the complex plastids visible in this photo.
Eukaryotic cells look very different from bacteria and archaea, with their more complex, compartmentalized structure; and in fact, if one examines some of their organelles — their mitochondria and chloroplasts — they will find much evidence to suggest that the eukaryotic cell is fundamentally different. Unlike the cells of bacteria and archaea, the eukaryotic cell appears to be a chimera of different cell types.
The eukaryotic cell as we know it is believed to have come into being when a cell (the ancestor to all modern eukaryotes) acquired a bacterial endosymbiont (an alphaproteobacterium). Over time, the bacterial endosymbiont evolved into the organelle known as the mitochondrion. A second endosymbiosis event is believed to have occurred much later, when one of these new chimeric cell types engulfed a photosynthetic cell (a cyanobacterium), which survived inside the host cell without being digested. The entrapped cyanobacterium became what is now known as the chloroplast, with the lineage of cells containing this new endosymbiont giving rise to the algae and land plants.
The endosymbiotic theory described above is well supported by scientific data. Evidence in support of this theory includes the following:
As is true for all cellular life forms, both mitochondria and chloroplasts have their own DNA and ribosomes. DNA in these organelles is typically present in a circular chromosome, as is normally the case in bacteria.
Various lines of evidence (e.g., ribosomal RNA sequence data, cytochrome c sequence data, whole genome sequences, protein content, gene clusters, genome organization) indicate that mitochondria and chloroplasts are descended from bacteria (e.g., Bonen and Doolittle 1975, Yang et al. 1985, Lang et al. 1997). Based on molecular phylogenies, the cell that gave rise to the mitochondrion was apparently a bacterium belonging to the class Alphaproteobacteria (Yang et al. 1985, Williams et al. 2007), while chloroplasts (and other plastids, which are derived from the chloroplast) apparently descended from cyanobacteria (Delwiche and Palmer 1996, Stoebe and Kowallik 1999).
Chloroplasts are structurally, biochemically, and functionally similar to cyanobacteria. Both chloroplasts and cyanobacteria are surrounded by two membranes. Within their interiors, they contain flattened membrane-bound sacs called thylakoids, where the light-dependent reactions of photosynthesis take place. Only chloroplasts and cyanobacteria are known to possess thylakoids, carry out oxidative photosynthesis, and have two photosystems (Photosystems I and II) (Raven and Allen 2003).
Mitochondria also share similarities with their ancestral counterparts. Both mitochondria and alphaproteobacteria are surrounded by two membranes. In mitochondria, the inner membrane has many folds in it, called cristae. The cristae increase the surface area of the membrane (where the electron transport chain reactions of respiration take place), which helps to maximize the amount of ATP that can be produced. Recent studies indicate that the mechanism involved in cristae formation was likely inherited from an alphaproteobacterial ancestor (Muñoz-Gómez et al. 2015, 2017).
As described by Muñoz-Gómez et al. (2015), the formation of cristae in the mitochondria of animals and fungi is controlled by a multiprotein complex called MICOS (mitochondrial contact site and cristae organizing system). The authors found homologs for at least two of the components of this complex—Mic60 and Mic10— in all major groups of eukaryotes, indicating that this mechanism of cristae formation is likely an ancient trait shared by all eukaryotes. When searching protein databases for bacteria and archaea, the authors found homologs of Mic60 (the central and largest protein of the complex) only in alphaproteobacteria. Some alphaproteobacteria possess intracytoplasmic membranes (ICMs) with morphologies that resemble mitochondrial cristae. Functionally, these ICMs provide the same benefit as cristae— they increase the surface area of the membrane for energy transduction processes (Niederman 2006). In addition, the versions of MICOS in mitochondria and alphaproteobacteria share sequence and structural similarities, and mitochondrial cristae and alphaproteobacterial ICMs develop similiarly (Muñoz-Gómez et al. 2017). These findings lend support to other molecular data that indicate mitochondria likely evolved from an alphaproteobacterium.
Because mitochondria and chloroplasts are derived from bacteria, many antibiotics that target bacteria adversely affect these organelles (Wang et al. 2015, Raven et al. 1992, Hong et al. 2015, Kalghatgi et al. 2013). For example, many antibiotics target bacterial ribosomes, binding to them at specific locations to inhibit protein synthesis (Böttger et al. 2001); however, the ribosomes of mitochondria and chloroplasts share many amino acid and nucleotide sequences with those of bacteria, so such antibiotics tend to also inhibit protein synthesis in these organelles (while eukaryotic ribosomes are unaffected) (Wang et al. 2015, Raven et al. 1992, Hong et al. 2015).
Mitochondria and chloroplasts are considered to be semi-autonomous organelles—as you may expect from entities that were once free-living—they carry out some of their own processes independent of the host cell in which they reside. For example, they encode and manufacture some of their own proteins, and they typically replicate their DNA out of phase with DNA replication of the host cell (i.e., not necessarily during the S phase of the host's cell cycle but at any random time) (Alberts et al. 1994). However, due to a phenomenon known as endosymbiotic gene transfer (a special type of horizontal gene transfer), these once free-living entities are now largely controlled by the host cell.
Endosymbiotic gene transfer is a continuous and naturally-occurring process whereby DNA is transferred from an organelle (or endosymbiont) to the nucleus of the host cell (Timmis et al. 2004). Many genes with distinct bacterial signatures have been found in the eukaryotic nucleus that apparently originated from the mitochondria and chloroplasts (Martin et al. 2002, Gray 2012). In addition, endosymbiotic gene transfer has been demonstrated and measured in laboratory experiments involving mitochondria in yeast (Thorsness and Fox 1990) and chloroplasts in tobacco plants (Huang et al. 2003). Endosymbiotic gene transfer may occur at a surprisingly fast rate and the transfer of genes is much more likely to occur from organelle to nucleus than in the reverse direction (Huang et al. 2003, Thorsness and Fox 1990). The large majority of the proteins necessary for the functioning of mitochondria and chloroplasts are encoded by genes that are now found in the nucleus. These proteins are manufactured using host cell ribosomes and then are imported back into the organelle (Gray 2012, Martin 2003).
Mitochondria and chloroplasts have highly reduced genomes compared to the genomes of their free-living counterparts. An estimated 0.5-1.2% of ancestral genes have been retained in the mitochondrial genome (Burger et al. 2013) while roughly 4-8% have been retained in the chloroplast genome (Huang et al. 2003; with number of genes for the most gene-rich plastids obtained from Janouškovec et al. 2013). Many genes no longer needed for survival have been lost entirely. Other genes, as already noted, have been transferred from these organelles to the nucleus. Although some of these transferred genes encode proteins that get imported back into the organelle, many of the genes that get transferred to the nucleus from organelles get repurposed—the proteins they encode are used elsewhere in the cell (Martin et al. 2002).
The chloroplasts of some photosynthetic eukaryotes, including glaucophyte algae and the moss Physcomitrella patens, still contain a peptidoglycan cell wall (Keeling 2004, Hirano et al. 2016), a remnant from their bacterial ancestor. The vast majority of bacteria, including cyanobacteria, have a cell wall, and the characteristic component of this wall—which is not found in the cell walls of eukaryotes or archaea— is peptidoglycan. In both cyanobacteria and the chloroplasts of glaucophyte algae (and perhaps in the chloroplasts of some other eukaryotes) the layer of peptidoglycan is found between the inner and outer membranes (Miyagishima et al. 2014).
Like bacteria, mitochondria and chloroplasts divide by binary fission, and some of the components involved in the division of these organelles are clearly of bacterial origin. These components, which include FtsZ proteins (found in chloroplasts and some mitochondria); and Min proteins, ARC6, peptidoglycan, and DipM (found in chloroplasts), play an important role in cell division in bacteria (Margolin 2005, Vitha et al. 2001, Miyagishima 2011, Miyagishima et al. 2014) but are not involved in the division of eukaryotic cells. Genes for the synthesis of these organelle components are found in the host cell nucleus (Miyagishima et al. 2014, Margolin 2005, Jackson et al. 2015)—apparent examples of endosymbiotic gene transfer. Two of these components will be elaborated further here: FtsZ and peptidoglycan.
The cytoskeletal protein FtsZ is of key importance in the cell division of most bacteria, including cyanobacteria and alphaproteobacteria (Arimura and Tsutsumi 2002). FtsZ is the main component of a contractile ring (called a Z-ring) that forms at the mid cell division site (Margolin 2005). This bacterial protein is involved in the division of mitochondria in many protoctists, and in the division of chloroplasts and related plastids in all chloroplast/plastid-containing organisms (except apicomplexans [Striepen et al. 2000]) (Margolin 2005, Vitha et al. 2001). The FtsZ protein found in mitochondria is different from that found in chloroplasts—when compared to the FtsZs of major groups of bacteria, the FtsZs in mitochondria are most closely related to that of alphaproteobacteria, while the FtsZs in chloroplasts are most closely related to that of cyanobacteria (Beech et al. 2000). Although FtsZs have been retained in the mitochondria of protoctists, they have been lost from the mitochondria of more complex organisms (i.e., animals, fungi and higher plants), where FtsZ has been replaced by eukaryotic dynamin-like proteins (Margolin 2005, Vitha et al. 2001). The host-cell-derived dynamin-like proteins are involved in the division of all mitochondria, even those that utilize FtsZ; they also play a role in the division of chloroplasts, so the organelle division apparatus often consists of a mixture of bacterial and eukaryotic components (Margolin 2005).
In bacteria, including cyanobacteria, peptidoglycan plays an important role in cell division. Although, peptidoglycan appears to have been lost from the chloroplasts of most algae and land plants, in many cases where it has been retained, it is an important component of the division apparatus (Machida et al. 2006). Antibiotics that target peptidoglycan, such as penicillin and ampicillin, inhibit chloroplast division in peptidoglycan-containing chloroplasts, such as those in the moss Physcomitrella patens (Katayama et al. 2003) and in glaucophyte algae (Berenguer et al. 1987). Similarly, knocking out genes involved in peptidoglycan biosynthesis affects the ability of peptidoglycan-containing chloroplasts to divide (Hirano et al. 2016). In both cases a similar effect results— the appearance of giant chloroplasts (Machida et al. 2006, Hirano et al. 2016). In addition to affecting the chloroplasts of moss and glaucophyte algae, treatment with peptidoglycan-targeting antibiotics has also been found to cause giant chloroplasts in a charophycean green alga, a liverwort, and a lycophyte, suggesting that peptidoglycan also plays a role in chloroplast division in these organisms (Hirano et al. 2016, Machida et al. 2006).[4] At least in the case of glaucophyte algae, the retention of peptidoglycan in the chloroplast has resulted in the preservation of a chloroplast division process that only involves bacterial division components (i.e., with no division components of host eukaryotic origin [Miyagishima et al. 2014]).
Organelles evolve at different rates in different types of organisms, and different genes may be lost depending on the organism (as evident from the presence of some bacterial components such as FtsZ and peptidoglycan in the organelles of some organisms but their absense from organelles of others). So, by looking at the gene and protein content of an organelle across a large range of taxa, scientists can obtain a better picture of which genes were likely present in the free-living ancestor. Based on various lines of evidence, scientists believe that all mitochondria are descended from a single endosymbiosis event (Gray 2001, 2012). A similar conclusion has been made with regards to chloroplasts (primary plastids) (Moreira et al. 2000, Archibald 2015, Keeling 2010). Although in chloroplasts, a more complex scenario may have occurred, where transient endosymbionts contributed genes to the nucleus (via endosymbiotic gene transfer), which ultimately affected the endosymbiont that became the chloroplast (Howe et al. 2008).
Scientists have identified other organelles in eukaryotes that apparently evolved from more recent endosymbiosis events. These examples provide even more convincing evidence that organelles can evolve from endosymbioses and that this phenomenon is not exceedingly rare but can occur repeatedly over evolutionary time.
One such example involves chloroplast-like organelles (originally named chromatophores) discovered in the amoeba Paulinella chromatophora. The endosymbiotic event giving rise to the photosynthetic organelle of this species is estimated to have occurred approximately 90-140 million years ago (Delay et al. 2016). In comparison, recent dating methods suggest that the endosymbiotic events associated with the chloroplast and mitochondrion took place approximately 900 million years ago and 1,200 million years ago, respectively (Shih and Matzke 2013). The plastid in Paulinella evolved from a different type of cyanobacterium than the one that gave rise to the chloroplast, and it evolved recently enough that scientists have been able to narrow down the identity of its ancestor—it was closely related to a clade consisting of Prochlorococcus and Synechococcus (Marin et al. 2005).
The same processes involved in the evolution of the mitochondrion and chloroplast have shaped the evolution of the "chromatophore"—i.e., gene loss, transfer of genes from the organelle to the nucleus, and the evolution of a mechanism to target essential proteins now encoded in the nucleus for import back into the organelle (Archibald 2015, Nowack et al. 2011). Paulinella's "chromatophore" is likely still evolving. It's genome contains 26% of the total number of genes found in its closely related free-living ancestor—Synechococcus (Nowack et al. 2008) (as compared to 4-8% of ancestral genes retained in the chloroplast genome, as noted earlier).

Primary Endosymbiosis. A heterotrophic eukaryotic cell engulfs a cyanobacterium, with the cell membrane of the heterotrophic cell wrapping around the cyanobacterium and then pinching off to form a phagosome. For some reason the cyanobacterium is not digested, and the phagosomal membrane is lost. The entrapped cyanobacterium evolves to become an organelle—the chloroplast (a primary plastid). The host cell acquires the ability to photosynthesize, becoming an alga.

Primary Endosymbiosis. A heterotrophic eukaryotic cell engulfs a cyanobacterium, with the cell membrane of the heterotrophic cell wrapping around the cyanobacterium and then pinching off to form a phagosome. For some reason the cyanobacterium is not digested, and the phagosomal membrane is lost. The entrapped cyanobacterium evolves to become an organelle—the chloroplast (a primary plastid). The host cell acquires the ability to photosynthesize, becoming an alga.

Primary Endosymbiosis. A heterotrophic eukaryotic cell engulfs a cyanobacterium, with the cell membrane of the heterotrophic cell wrapping around the cyanobacterium and then pinching off to form a phagosome. For some reason the cyanobacterium is not digested, and the phagosomal membrane is lost. The entrapped cyanobacterium evolves to become an organelle—the chloroplast (a primary plastid). The host cell acquires the ability to photosynthesize, becoming an alga.
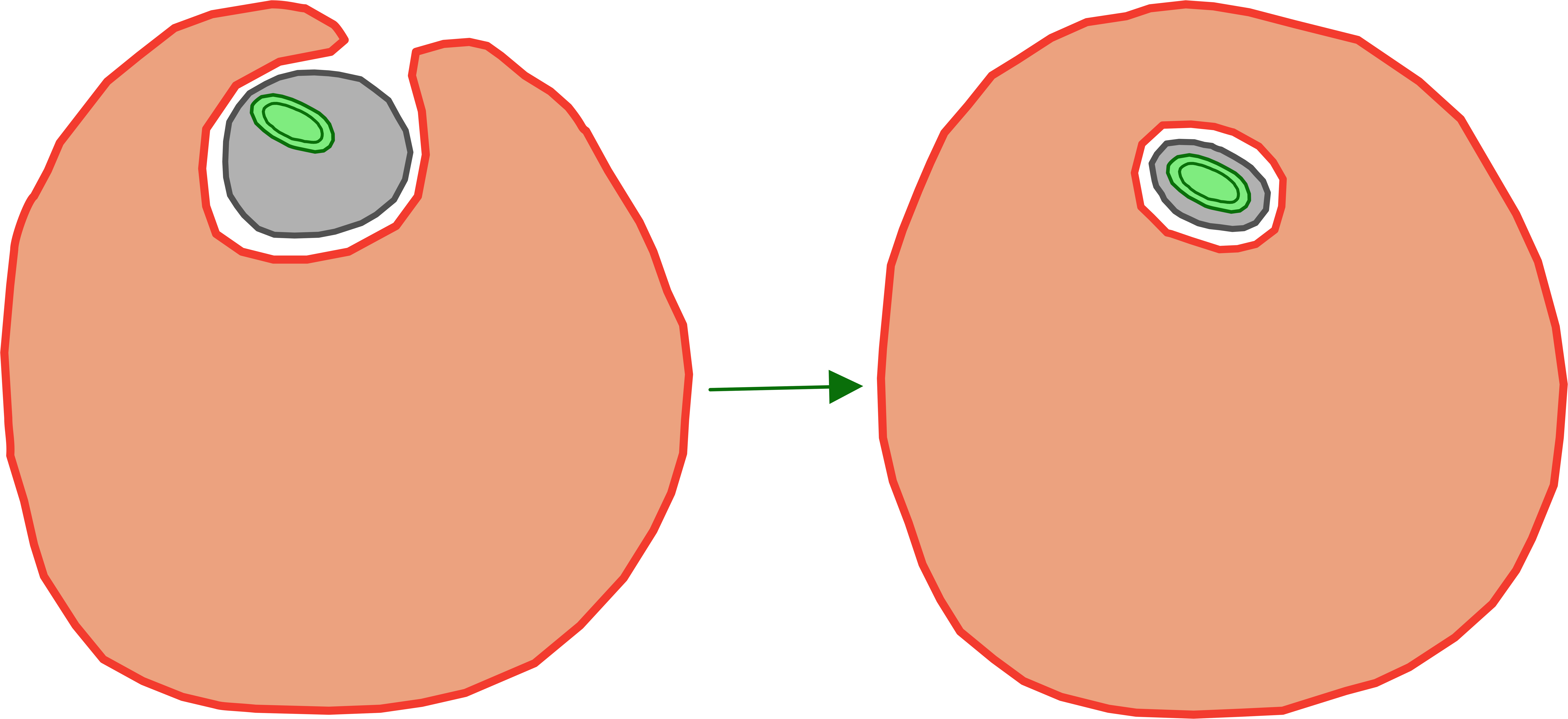
Secondary Endosymbiosis. A heterotrophic eukaryotic cell engulfs a photosynthetic eukaryote (an alga with a chloroplast). The resulting organelle, called a complex plastid or secondary plastid, is surrounded by four membranes. The two innermost membranes (shown within and along the outside edge of the green area) correspond to the membranes of the cyanobacterium/chloroplast. The membrane exterior to that (in dark gray) corresponds to the cell membrane of the organism that engulfed the cyanobacterium. The outermost membrane (in red) is derived from the phagosomal membrane of the host cell (Keeling 2010). In some algae with secondary plastids, a vestigial nucleus (called a nucleomorph) is found between the second and third membranes of this organelle (Keeling 2004) (within the lighter gray area shown).

Secondary Endosymbiosis. A heterotrophic eukaryotic cell engulfs a photosynthetic eukaryote (an alga with a chloroplast). The resulting organelle, called a complex plastid or secondary plastid, is surrounded by four membranes. The two innermost membranes (shown within and along the outside edge of the green area) correspond to the membranes of the cyanobacterium/chloroplast. The membrane exterior to that (in dark gray) corresponds to the cell membrane of the organism that engulfed the cyanobacterium. The outermost membrane (in red) is derived from the phagosomal membrane of the host cell (Keeling 2010). In some algae with secondary plastids, a vestigial nucleus (called a nucleomorph) is found between the second and third membranes of this organelle (Keeling 2004) (within the lighter gray area shown).

Secondary Endosymbiosis. A heterotrophic eukaryotic cell engulfs a photosynthetic eukaryote (an alga with a chloroplast). The resulting organelle, called a complex plastid or secondary plastid, is surrounded by four membranes. The two innermost membranes (shown within and along the outside edge of the green area) correspond to the membranes of the cyanobacterium/chloroplast. The membrane exterior to that (in dark gray) corresponds to the cell membrane of the organism that engulfed the cyanobacterium. The outermost membrane (in red) is derived from the phagosomal membrane of the host cell (Keeling 2010). In some algae with secondary plastids, a vestigial nucleus (called a nucleomorph) is found between the second and third membranes of this organelle (Keeling 2004) (within the lighter gray area shown).
Other more recent endosymbiotic events include those that gave rise to the so-called "complex" plastids. A plastid is a photosynthetic organelle or an organelle that evolved from a photosynthetic organelle. Plastids may be referred to as "primary" (as in the chloroplast and nonphotosynthetic plastids evolved from the chloroplast [also the "chromatophores" of Paulinella chromatophora]; these arose as the result of a single endosymbiosis event), "secondary" (arising from two sequential endosymbiosis events), and even "tertiary" (arising from three sequential endosymbiosis events) (Howe et al. 2008). Secondary and tertiary plastids are considered to be complex plastids.
Primary plastids, as already described, are evolved from a cyanobacterium that was engulfed (through the process of phagocytosis) by a heterotrophic eukaryote. Phagocytosis results in the formation of a membrane-bound compartment, called a phagosome, which contains the engulfed food item. The membrane surrounding the phagosome is derived from the part of the cell membrane that was pinched off to form the compartment. Normally the phagosome fuses with a vesicle that contains enzymes (a lysosome) and the food item is digested. But in this case, digestion failed to occur, and the cyanobacterium became an endosymbiont within the host cell. As the endosymbiont transitioned into an organelle, many of its genes were lost and others were transferred to the host cell's nucleus through the process of endosymbiotic gene transfer. Primary plastids are surrounded by two membranes, which correspond to the inner and outer membranes of the cyanobacterium; the phagosomal membrane surrounding the cyanobacterium was apparently lost (Keeling 2004). Primary plastids are found in red algae, glaucophyte algae, green algae, and land plants (Miyagishima 2011).
Secondary plastids are surrounded by four (sometimes three) membranes (Keeling 2004, Howe et al. 2008). They came into being when a eukaryotic cell containing a chloroplast (primary plastid) was engulfed by a larger, nonphotosynthetic eukaryotic cell. For some reason the engulfed cell was not digested but remained entrapped within the host cell. Over time, many genes from the nucleus of the engulfed cell were lost, and some of its other genes were transferred to the nucleus of the host cell (Gilson et al. 1997, Keeling 2004). Some genes from the engulfed cell's chloroplast may also have been transferred to the nucleus of the host cell (Keeling 2004). In most lineages of algae that contain secondary plastids, the nucleus of the engulfed cell was eventually lost. However, in cryptophytes and chlorarachniophytes a vestigial nucleus remains (along with eukaryotic ribosomes and a small amount of cytoplasm [Ludwig and Gibbs 1989, Douglas et al. 2001]), sandwiched between membranes of the complex plastid (Gilson et al. 1997). Secondary endosymbiotic events apparently occurred on at least three separate occasions, probably more (Gilson et al. 1997, Rogers et al. 2007). In all known cases, either a red or green alga was engulfed (Keeling 2004). Taxa containing secondary plastids that are derived from red algae include cryptophytes, heterokonts (stramenopiles), haptophytes, apicomplexa and dinoflagellates; those with secondary plastids derived from green algae include chlorarachniophytes, euglenids and some dinoflagellates (Keeling 2004, Rogers et al. 2007, Keeling 2004).
Other plastids are believed to have evolved as a result of even more complex scenarios—e.g., via serial secondary endosymbiosis or tertiary endosymbiosis. In serial secondary endosymbiosis, an alga containing a secondary plastid loses its plastid, but then later acquires a new secondary plastid (by obtaining another alga endosymbiont with a primary plastid). Serial secondary endosymbiosis is believed to have occurred in the dinoflagellate Lepidodinium (Keeling 2004), and possibly in diatoms (Burki et al. 2014). In tertiary endosymbiosis, a eukaryotic cell engulfs an alga with a secondary plastid, with the engulfed alga evolving into a tertiary plastid. An interesting example of a tertiary plastid occurs in a type of dinoflagellate known as a "dinotom." The plastid in dinotoms was formerly an entrapped diatom. Although the plastid no longer looks like a diatom because it lost its cell wall (Imanian and Keeling 2007), it has retained ample evidence of its diatom ancestry, including a nucleus (with many genes still intact), cytoplasm, ribosomes, plastids, endoplasmic reticulum, and mitochondria (Burki et al. 2014, Imanian and Keeling 2007).
Open Questions Regarding the Origin of Eukaryotic Cells
Today there is broad agreement among scientists that mitochondria and plastids evolved from ancient endosymbiotic events. However, scientists don't agree on all of the details— especially with regards to the origin of the mitochondrion. In particular, they don't agree on the type of cell that gave rise to the mitochondrion or the type of cell that became the host in this endosymbiosis.
Nature of the Mitochondrial Endosymbiont
Although the endosymbiont that gave rise to the mitochondrion was apparently an alphaproteobacterium, theories vary on whether that cell was photoautotrophic or heterotrophic; obligately aerobic or facultatively anaerobic (Martin et al. 2015, Kurland and Andersson 2000, Cavalier-Smith 2006). Consequently, there is a lack of agreement over what the original benefit was that the endosymbiont provided to the host cell.
Today in most eukaryotes the mitochondrion is the site where aerobic respiration takes place— it is the location in the cell where the vast majority of ATP is produced. Most of that ATP is generated through use of an electron transport chain located in the inner membrane of the mitochondrion. In essence, in the process of respiration, high energy electrons are extracted from food and are fed into an electron transport chain. The chemical reactions of the electron transport chain release energy from the electrons, a bit at a time, and some of that energy is captured and stored in the form of transmembrane proton gradient. The force generated by this gradient is harnessed to produce ATP. This process is explained in more detail in the box below.
With one known exception, all eukaryotes have some form of a mitochondrion (i.e., either a mitochondrion or an organelle derived from the mitochondrion or protomitochondrion). The exception is the oxymonad Monocercomonoides sp., which apparently lacks any sign of such an organelle (Karnkowska et al. 2016). Oxymonads are single-celled anaerobic protoctists that have flagella and are known to live only within the guts of animals (Heiss and Keeling 2006). Although Monocercomonoides sp. apparently lacks a mitochondrial organelle, its closely related sister taxon Paratrimastix has a mitochondrion— or rather a highly reduced form of a mitochondrion. Given the presence of this organelle in its sister taxon, it is likely that Monocercomonoides sp. evolved from an ancestor that had a mitochondrion, but over time the organelle ceased to provide a vital function in the cell and was eventually lost (Karnkowska et al. 2016).
Aerobic eukaryotes have a typical mitochondrion— an aerobic mitochondrion, which generates ATP via aerobic respiration. In eukaryotes that live in low oxygen environments, the mitochondrial organelles tend to be much reduced in size and function (Shiflett and Johnson 2010). They are not used for aerobic respiration, and in most cases, they're not used for anaerobic respiration either. Rather than being called "mitochondria" they are generally referred to by names such as "organelles of mitochondrial origin" (Müller et al. 2012) or "mitochondrion-related organelles" (MROs) (Stairs et al. 2015). Like mitochondria, MROs are surrounded by a double membrane. In some taxa, some of the mitochondrial DNA has been retained—and it is homologous to DNA found in aerobic mitochondria (Gray 2012). In other taxa all genes have been lost from the organelle. However, even the organelles lacking genetic material can be traced back to mitochondria by the presence of mitochondrial marker proteins— proteins found in the organelle that are of distinct mitochondrial origin (Karnkowska et al. 2016, Gray 2012).
Eukaryotes living in low oxygen environments were once described as "amitochondriate"— because initially it was not known that they possessed mitochondrial organelles— because the organelles were so highly reduced (Shiftlett and Johnson 2010). Given their apparent "lack" of mitochondria, scientists believed that these organisms belonged to a distinct and primitive lineage of eukaryotes— a lineage that never inherited the mitochondrion. Essentially they were thought to be living representatives of ancient eukaryotes or "archezoans." However, it is now known that these so-called "archezoans" don't form a phylogenetic group. They are widely scattered across the divergent phylogenetic branches of the eukaryotes, and in all cases they apparently evolved from ancestors containing bona fide mitochondria (Stairs et al. 2015, Shiftlett and Johnson 2010, Müller et al. 2012).
Thus available evidence indicates that
- the protoeukaryotic cell acquired an alphaproteobacterial endosymbiont, which evolved into the mitochondrion, and
- over time, the mitochondrion-containing progeny of the protoeukaryotic cell expanded into new environments and diversified, forming the different lineages of eukaryotes that are present today, and
- in many of those lineages, some of the taxa became adapted to
living in areas with low oxygen concentrations and, in some cases, locations with plentiful
food supplies (e.g. within the guts of animals). In these taxa, the mitochondrion
became reduced in size and function due to one or more of the following reasons:
- because aerobic respiration was no longer possible in such environments, and/or
- because an efficient energy metabolism was not needed (or favored) due to an ample food supply, and/or
- because the organism acquired new adaptations— metabolic processes that could be carried out elsewhere in the cell, which replaced crucial functions that were previously performed by the mitochondrion.
So, in general, in a given lineage of eukaryotes, organisms living independent lifestyles in oxygen-rich environments retained high functioning mitochondria, while other organisms from the same lineage— those that became adapted to low oxygen environments— had mitochondria that became diminished over time, as functions were no longer needed.
Mitochondria and related organelles have been classified based on their different functions (Müller et al. 2012)—however, it should be noted that some MROS defy simple classification, as they appear to have characteristics that span multiple categories of mitochondrial organelles (Maguire and Richards 2014). Some described types of mitochondria and MROs (from Müller et al. 2012) include the following:[5]
- Aerobic mitochondria (Produce ATP by aerobic respiration)
- Anaerobic mitochondria (Generate ATP by means of anaerobic respiration and don't produce H2)
- H2-producing mitochondria (Generate ATP and produce H2 and have an electron transport chain. Although a proton gradient is produced in the mitochondrion, possibly by the electron transport chain, ATP is not believed to be synthesized from this gradient (due to an apparent lack of ATP synthase) (Müller et al. 2012). Hence ATP is likely produced solely by means of substrate level phosphorylation as is the case in hydrogenosomes. (This type of MRO is considered to be a "missing link" between mitochondria and hydrogenosomes [Boxma et al. 2005, Martin 2005])
- Hydrogenosomes (Generate ATP and produce H2 via fermentations; do not have an electron transport chain)
- Mitosomes (Do not generate ATP. Organisms containing mitosomes tend to be parasites, and ATP is produced in the cytosol via substrate level phosphorylation or is obtained directly from the host's cells [Shiftlet and Johnson 2010, Tsaousis et al. 2008])
Although mitochondrial organelles vary in their means of ATP production (or lack thereof, in the case of the mitosomes), these organelles tend to have at least one crucial function in common— the synthesis of iron-sulfur clusters via the iron-sulfur cluster (ISC) assembly system (Shiftlet and Johnson 2010, Karnkowska et al. 2016, Maguire and Richards 2014). Iron-sulfur clusters are small molecules that are associated with many different proteins and are involved in a number of essential biochemical reactions in cells (Lill and Mühlenhoff 2008). For example, some iron-sulfur clusters bind to enzymes to activate them, others may serve as electron carriers in electron transport chains. The ISC assembly system is of bacterial origin and it was apparently inherited by eukaryotes from the mitochondrial endosymbiont (Lill and Mühlenhoff 2008). Using the ISC assembly system, mitochondria manufacture their own iron-sulfur proteins, and they begin the manufacturing process for iron-sulfur proteins that are used in the cytosol and nucleus (Kaut et al. 2000, Lill and Mühlenhoff 2008).[6]
The few known taxa having MROs that lack the ISC assembly system, have replaced this system with an alternate bacterial system, which was apparently obtained through horizontal gene transfer (Karnkowska et al. 2016). In fact, the one eukaryote that appears to be missing a mitochondrial organelle, Monocercomonoides sp., lacks the ISC assembly system but possess genes for an alternative, analogous system— the sulfur mobilization (SUF) system (Karnkowska et al. 2016). Monocercomonoides' closely related sister taxon Paratrimastix pyriformis, which has an MRO (Karnkowska et al. 2016), also lacks an ISC assembly system (Karnkowska and Hampl 2016) but contains an SUF system (Karnkowska et al. 2016). In both taxa, the SUF components were found to lack mitochondrial targeting signals, indicating that the system was apparently not localized to a mitochondrial organelle (Karnkowska et al. 2016). In Monocercomonoides, the SUF system was found to be localized in the cytosol (localization of the SUF system in Paratrimastix pyriformis was not determined [Karnkowska and Hampl 2016]). Based on results of a phylogenetic analysis, Monocercomonoides and Paratrimastix apparently inherited the SUF system from a common ancestor, which acquired it through horizontal gene transfer from a bacterium. The authors proposed that sometime after the two lineages diverged, the Monocercomonoides lineage lost its MRO because the ISC system in the MRO was replaced by the cytosolic SUF system and presumably the MRO no longer had a crucial function in the cell (Karnkowska et al. 2016). Paratrimastix however, retained its MRO (Karnkowska et al. 2016, Zubáčová et al. 2013, Karnkowska and Hampl 2016 ), in all likelihood because its MRO continues to carry out some indispensible function. The function of Paratrimastix's MRO is not yet well known, but at a minimum is believed to include amino acid metabolism (Zubáčová et al. 2013, Karnkowska and Hampl 2016).
✦✦✦
To obtain a clearer picture of the nature of the endosymbiont that gave rise to the mitochondrion, one can look at the genes present in the most gene-rich mitochondria known— that of the jakobid flagellates (Burger et al. 2013). The mitochondrial genome of the jakobids is the most bacteria-like mitochondrial genome known and appears to have diverged the least from that of its endosymbiotic ancestor (Lang et al. 1997). The jakobids are single-celled biflagellate organisms, which for the most part contain typical aerobic mitochondria (Burger et al. 2013, Leger et al. 2016).[7] The mitochondrial DNA of the jakobids includes genes involved in aerobic respiration, including those associated with electron transport and ATP synthesis (Burger et al. 2013).
Another approach that has been taken to determine the nature of the mitochondrial endosymbiont has involved identifying genes in a eukaryote's nucleus that are of apparent alphaproteobacterial ancestry (excluding any genes showing evidence of recent horizontal gene transfer), and assuming that these genes originated from the endosymbiont (Gray 2012). In addition, scientists have attempted to reconstruct the proteome (collective set of proteins) of the endosymbiont by identifying eukaryotic proteins that are apparently closely related to those of alphaproteobacteria, and assuming that these were present in the mitochondrial endosymbiont (Gray 2015). Based on data from these types of studies, the mitochondrial endosymbiont contained pathways for the synthesis of lipids, biotin, heme, and iron-sulfur clusters and apparently had the ability to respire aerobically (Gray 2012, 2015; Szklarczyk and Huynen 2010).
So we know that the endosymbiont was an alphaproteobacterium, that it had an electron transport chain, and that it was capable of aerobic respiration. But we don't know whether it was an obligate aerobe, i.e., required oxygen to survive, or whether it was capable of living an anaerobic lifestyle, generating ATP by fermentations, but respiring aerobically when oxygen became available. We also don't know whether the endosymbiont was able to photosynthesize. There are in fact some bacteria (e.g. some non-sulfur purple bacteria, which are a type of alphaproteobacteria) that carry out both anaerobic photosynthesis and aerobic respiration—using the same electron transport chains (Fenchel and Finlay 1995). Many theories have been proposed about the nature of the endosymbiont's metabolism—covering a range of possibilities— but they are all, at least at this point in time, merely speculative.
Nature of the Host Cell that Acquired the Mitochondrial Endosymbiont
Scientists have also speculated about the metabolism of the host cell, describing it, in wildly differing theories, as heterotrophic or autotrophic, obligately anaerobic or capable of aerobic respiration (Martin et al. 2015). Unfortunately trying to determine the original metabolic workings of the host cell is even more difficult than it is for the endosymbiont. At least in the case of the mitochondrion, researchers have been able to gain clues to its past by looking at the genes in the vestigial genome of the organelle and by identifying genes of alphaproteobacterial ancestry in the host's nucleus. However, there isn't an easy way to determine the original processes and functions of the host cell from looking at the current components and functions of eukaryotic cells. You could identify proteins that are found only in eukaryotes (Hartman and Fedorov 2002), which could provide information about cellular processes, but determining whether these proteins were present in the host cell prior to the endosymbiosis event (or whether they evolved or were acquired later) is another matter. It is perhaps unlikely that we will ever be able to confirm or refute some of the hypotheses regarding the original metabolisms and metabolic exchanges between the host and endosymbiont.
A bigger question regarding the host cell has concerned its evolutionary origin. At the most fundamental level, scientists have been trying to determine what type of cell it was— what cellular domain it belonged to. While researchers have been able to amass a great deal of evidence indicating that the mitochondrial endosymbiont was a bacterium, determining the nature of the host cell has proven to be much more challenging. There are two viewpoints that have been commonly argued: (1) that the host cell was an archaeon, which evolved to become a eukaryote only after acquiring the alphaproteobacterial endosymbiont (i.e., there were originally only two cellular domains—archaea and bacteria— and eukaryotes came into being as a result of a fusion of those two domains); and (2) that the host cell, at the time that it acquired the endosymbiont, was a eukaryote or at least a simpler version of a eukaryote (i.e., there were already three distinct cellular domains prior to the host cell acquiring the proto-mitochondrion).
The ribosomal RNA sequencing performed by Carl Woese and colleagues (Woese et al. 1990) showed eukaryotes to be slightly more related to archaea than bacteria, but they noted that eukaryotes and archaea "are sufficiently dissimiliar, and they diverged so early, that little would be gained by defining a taxon that encompasses both." Thus they proposed that Bacteria, Archaea, and "Eucarya" should be recognized as three distinct cellular domains. In Woese's tree of life, the Archaea and "Eucarya" are sister taxa— they are two different groups that share a common ancestor (which in this case is not shared by bacteria).
Other scientists performing phylogenetic analyses— in some cases using the same data as Woese— have come up with different trees— ones in which eukaryotes form a group that is located within the Archaea (Lake 1988, Cox et al. 2008, Foster et al. 2009, Williams et al. 2012). Thus these trees recognize only two cellular domains— Bacteria and Archaea.
At the time of Woese's work, archaea had been only newly discovered and had not been extensively sampled. To identify them, they had to be grown in culture; however archaea are notoriously difficult to culture in the laboratory— only a seemingly small percentage of taxa can be easily cultured, which had made it impossible to determine the extent of their distribution and abundance. This situation changed, when new molecular biology techniques became available that enabled researchers to analyze DNA and identify genomes directly from environmental samples without having to isolate and culture organisms. Using these culture-free techniques scientists have since discovered some archaea that appear to be very closely related to eukaryotes (Spang et al. 2015, Seitz et al. 2016, Zaremba-Niedzwiedzka et al. 2017).
The most eukaryote-like organisms identified to date include the so-called Lokiarchaeota, Thorarchaeota, Odinarchaeota, and Heimdallarchaeota phyla; together constituting a clade known as the Asgard superphylum (Zaremba-Niedzwiedzka et al. 2017, Spang et al. 2015). These archaea were identified from samples collected from deep marine sediments(Lokiarchaeota and Heimdallarchaeota), estuary sediments (Thorarchaeota), and hot springs (Odinarchaeota) (Spang et al. 2015, Seitz et al. 2016, Zaremba-Niedzwiedzka et al. 2017). Phylogenomic analyses performed by Zaremba-Niedzwiedzka et al. (2017) placed eukaryotes within the Asgard superphylum. In addition, many eukaryotic signature proteins— proteins once thought to occur only in eukaryotes— were identified in the Asgard genomes that were reconstructed from the samples.
Eukaryotic signature proteins include proteins associated with a complex internal membrane system, a complex internal signaling system, and a dynamic cytoskeleton, as well as other proteins involved in phagocytosis— a process known to occur only in eukaryotes (Hartman and Fedorov 2002).
Although recently, eukaryotic signature proteins have been found in some archaea (Spang et al. 2015), including archaea grown in culture (Ettema et al. 2011, Yutin and Koonin 2012, Lindås et al. 2008), to date, the greatest number of eukaryotic signature proteins (outside of eukaryotes) have been detected in the Asgard archaea (Zaremba-Niedzwiedzka et al. 2017). Also, based on analysis of the reconstructed Lokiarchaeota genome, the Asgard archaea appear to have the most eukaryote-like ribosome (Spang et al. 2015).
Another piece of evidence that appears to support the "two-domain" hypothesis has to do with the apparent chimeric origin of eukaryotic genes. Genes may be classified as "informational" or "operational." Informational genes include those involved in transcription, translation, and replication; their gene products have many molecular interactions in cells; while operational genes, known as "housekeeping genes," include genes for energy metabolism, biosynthesis pathways, and other processes important in cell maintenance; they tend to operate independently of other cellular components (Rivera et al. 1998, Jain et al. 1999, Boto 2010). Due to the different nature of these gene types, informational genes tend to be inherited vertically and are seldom transferred horizontally while operational genes are commonly transferred horizontally (Jain et al. 1999).
Genomic analyses performed by Rivera et al. (1998), found that in general, informational genes in eukaryotes are of archaeal origin, while operational genes in eukaryotes are primarily of bacterial descent. Since informational genes are mostly transmitted vertically, this result is consistent with the hypothesis that the protoeukaryotic cell was an archaeon.
Advocates of the two domain theory also point to the apparent absence of true archezoans— primitive eukaryotic cells— as providing further support for their theory. No truly amitochondriate lineage of eukaryotes has yet been discovered (Archibald 2015, Karnkowska et al. 2016). If the host cell was a eukaryote, then the descendents of the other eukaryotes living at that time, which didn't acquire the endosymbiont, must have all gone extinct— or be really difficult to find (Yutin et al. 2009, Gray 2012, Archibald 2015).
Given the number of recent studies showing the placement of eukaryotes within the Archaea, the two domain model is currently the most popular theory for explaining the evolution of eukaryotes. However, this theory leaves some lingering unanswered questions.
For example, it is not known how the endosymbiont ended up inside the host cell. No archaeon or bacterium alive today is known to be capable of phagocytosis (Yutin et al. 2009). Only eukaryotes have the ability to engulf other cells. However, there is at least one known example of a bacterial endosymbiont inside other bacterium—a gammaproteobacterium inside a betaproteobacterium, found within cells of mealybugs (Husnik and McCutcheon 2016). So such an endosymbiosis is clearly possible, but how it occurs has not been determined.
One explanation that has been proposed is that the endosymbiont was a predatory bacterium (Guerrero et al. 1986). Some bacteria are known to prey on other bacterial cells. They tend to be much smaller than the cells they attack and they are able to penetrate their prey/host cell, ending up in the cytoplasm or periplasm, where they then divide (Guerrero et al. 1986). Normally they degrade/consume the cytoplasm and kill the prey cell, although perhaps in some rare event the predator turned into a stable endosymbiont. This may have possibly occurred if the host cell provided the predator with a stable supply of nutrients, so that the predator did not need to directly consume the host (Davidov et al. 2006). Predatory bacteria capable of penetrating host cells include deltaproteobacteria, although it has been suggested that this predatory ability may have been present in ancestral forms of alphaproteobacteria (Davidov et al. 2006).
An alternative explanation—which appears to be gathering more support lately—is that the host cell had some ability to uptake small particles or macromolecules, and under rare circumstances it may have accidentally engulfed cells—including the alphaproteobacterium that became the mitochondrion. This idea is supported by the discovery of many eukaryotic signature proteins in the Asgard archaea— including proteins that are involved in phagocytosis in eukaryotes (Zaremba-Niedzwiedzka et al. 2017, Spang et al. 2015).
For a long time the process of endocytosis was believed to be restricted to eukaryotes. In endocytosis a cell uptakes material from the environment by forming a pocket in its membrane, which surrounds the material. The membrane at the open ends of the pocket then fuses forming an internal vesicle containing the material. Phagocytosis, which involves the uptake of large particles or cells is a type of endocytosis.
Recently scientists have documented an endocytosis-like process in a bacterium from the phylum Plantomycetes (Lonhienne et al. 2010). The bacterium was observed to uptake protein from the environment in an energy-dependent, receptor-mediated process, which involved invagination of the cytoplasmic membrane, and apparent formation of vesicles containing the internalized protein.
Given the presence of an endocytic process in a bacterium and the detection of many eukaryotic signature proteins in Asgard archaea, scientists have speculated that the archaeal protoeukaryotic cell could have had primative phagocytic abilities. The full expression of phagocytosis (and macropinocytosis), as seen in modern eukaryotes, involves the formation of membrane projections supported by actin filaments; in contrast, other forms of endocytosis involve only membrane invaginations (Yutin et al. 2009). There is some evidence to suggest that modern phagocytosis, which enables cells to engulf other cells, evolved relatively late and independently in different lineages of eukaryotes (Yutin et al. 2009). According to some scientists it is energetically impossible for a cell lacking mitochondria to have evolved full phagocytic abilities (Lane 2011). Thus many researchers believe that it is unlikely that the protoeukaryotic cell was capable of modern phagocytosis. However, it may have had limited phagocytic abilities, which on rare occasions enabled it to "accidentally" engulf cells, including the protomitochondrion (Yutin et al. 2009, Spang et al. 2015, Zaremba-Niedzwiedzka et al. 2017).
Another question that the two-domain theory has to contend with concerns the nature of the eukaryotic cell membrane. When cells divide, forming new cells, the plasma membrane of the daughter cells is never built from scratch, it is always inherited from the mother cell. In fact no cell at any stage in its life cycle has the ability to generate a new plasma membrane de novo (Beisson 2008, Harold 2010). Cell membranes are inherited through what is known as structural inheritance or non-DNA inheritance (Beisson 2008). A cell can repair its membrane, manufacture proteins to be inserted into the membrane, and also encode proteins (enzymes) for the biosynthesis of membrane components; it can even modify the amounts of certain lipids and/or other components so that the membrane remains functional under different environmental conditions (Siliakus et al. 2017); however, the overall structure of the membrane does not normally change.
A problem with the two-domain theory is that the cell membrane of archaea is fundamentally different from that of eukaryotes. Eukaryotes and bacteria have cell membranes that are chemically and structurally similar to one another but the archaeal membrane is quite different. The membrane lipids in Bacteria and Eukarya typically consist of a glycerol phosphate backbone connected via ester bonds to straight-chain fatty acid "tails" (Villanueva et al. 2017, Caforio et al. 2018). In contrast, in archaea, the glycerol phosphate backbone is linked via ether bonds to branched isoprenoid "tails" (Villanueva et al. 2017, Caforio et al. 2018). However, the most important difference between the two types of membrane lipids— i.e., the difference that appears to hold true without exception— is that the glycerol phosphate backbone in the archaeal cell membrane is the mirror image form of that found in the eukaryotic and bacterial cell membrane. (They are said to have opposite stereochemistry or to have different chirality or "handedness"; i.e., as in the case of right and left hands, the alternate glycerol phosphate backbones are mirror images of one another and they can't be rotated or shifted in any way so that they match up identically.) Archaeal membrane lipids are characterized by an sn-glycerol-1-phosphate (G1P) backbone whereas eukaryotes and bacteria have the mirror image sn-glycerol-3-phosphate (G3P) backbone (Villanueva et al. 2017, Caforio et al. 2018). These two different types of glycerol phosphate backbones are synthesized by different, evolutionarily unrelated enzymes (Koga and Morii 2007). All known eukaryotes and all known naturally-occurring bacteria have membrane lipids with the G3P glycerol phosphate backbone, whereas all known archaea have membrane lipids with the G1P glycerol phosphate backbone (Villanueva et al. 2017, Koga et al. 2003, Shimada and Yamagishi 2011). Given the apparent improbability of a change in membrane structure, it's not clear why eukaryotes—if descended from archaea— would have cell membranes with lipids that are bacteria-like rather than archaea-like.

Membrane lipids of bacteria and eukaryotes consist of an sn-glycerol-3-phosphate (G3P) "backbone" connected via ester linkages to two unbranched fatty acid "tails". Archaea have the mirror image sn-glycerol-1-phosphate (G1P) backbone, which is connected via ether linkages to two branched isoprenoid tails. Without exception, all known eukaryotes and all known naturally-occurring bacteria have membrane lipids with a G3P backbone, while all known archaea have membrane lipids with a G1P backbone.

Membrane lipids of bacteria and eukaryotes consist of an sn-glycerol-3-phosphate (G3P) "backbone" connected via ester linkages to two unbranched fatty acid "tails". Archaea have the mirror image sn-glycerol-1-phosphate (G1P) backbone, which is connected via ether linkages to two branched isoprenoid tails. Without exception, all known eukaryotes and all known naturally-occurring bacteria have membrane lipids with a G3P backbone, while all known archaea have membrane lipids with a G1P backbone.

Membrane lipids of bacteria and eukaryotes consist of an sn-glycerol-3-phosphate (G3P) "backbone" connected via ester linkages to two unbranched fatty acid "tails". Archaea have the mirror image sn-glycerol-1-phosphate (G1P) backbone, which is connected via ether linkages to two branched isoprenoid tails. Without exception, all known eukaryotes and all known naturally-occurring bacteria have membrane lipids with a G3P backbone, while all known archaea have membrane lipids with a G1P backbone.

In bacteria and eukaryotes, the cytoplasmic membrane is almost always arranged in a bilayer— the lipid molecules are in two layers with the hydrophilic glycerol phosphate backbones at the outside edges of the membrane and the hydrophobic tails in the interior of the membrane. Bilayer membranes are also found in some archaea.
Monolayer membranes are typically found in cells that occur at high temperatures and/or in extremely acidic environments (i.e., they are found in most archaea as well as in some thermophilic and acidophilic bacteria). The monolayer arrangement is believed to benefit cells under such conditions by making the membrane more stable and reducing its permeability. In a monolayer membrane, a single lipid molecule spans the membrane. A lipid in a monolayer membrane has a "backbone" at both ends of the molecule, with the "tail" portion in the middle. These membrane-spanning lipids are believed to be the result of a condensation reaction, which causes the tail ends of opposing lipids in a bilayer to fuse together (Siliakus et al. 2017).
For a cell to completely change its membrane structure— from archaeal-type lipids with a G1P glycerol phosphate backbone to bacterial-type lipids with a G3P glycerol phosphate backbone— the following would need to occur:
- the cell would need to receive—most likely through horizontal gene transfer— all of the genes necessary for the bacterial lipid biosynthesis pathway.
- the bacterial lipids would need to be manufactured and successfully incorporated into the membrane (without adversely affecting the cell's ability to survive or reproduce), and
- there would need to be some sort of selective mechanism, which would cause the bacterial lipids to be retained in the membrane and the archaeal lipids to be lost.
In addition to all of that, the cell would need to have or acquire a set of integral membrane proteins that are capable of functioning in the new membrane. Integral membrane proteins are proteins that are permanent components of cell membranes. They are "integral" to the structure and functioning of a membrane and they are involved in important biological processes that occur in cells. These proteins are specifically adapted to function in the physical and chemical environment of the particular phospholipid membrane in which they occur (Pogozheva et al. 2013). A change in the physicochemical nature of a membrane, such as that which would occur if an archaeal cell were to acquire a bacteria-type membrane, could cause an integral membrane protein to become nonfunctional.
Some scientists believe that the acquisition of a bacterial endosymbiont — the protomitochondrion— was the event that precipitated the change in the archaeal host cell's membrane (Lane 2015). Certainly, such an event would have provided the host cell with all the genes necessary to produce a bacterial-type membrane. But exactly how such a radical transformation would have occurred is unknown.
There is no known selective mechanism that would favor one mirror image form of the glycerol phosphate backbone over the other. This is because the chirality of the glycerol phosphate backbone does not affect the physicochemical properties of a membrane (Koga and Morii 2007). The type of linkage (ether vs ester) and type of hydrocarbon tail (isoprenoid vs fatty acid) do significantly change the properties of a membrane (Koga and Morii 2007). There are some bacteria with ether-linked membrane lipids (as summarized in Weijers et al. 2006), and some archaea with membranes containing fatty acid chains (Gattinger et al. 2002); however, the stereochemistry of the glycerol phosphate background that is characteristic of the domain is still maintained— that is, the bacteria have a G3P glycerol phosphate backbone and the archaea have the mirror image G1P backbone.
To explain the so-called "lipid divide"— the difference in the stereochemistry of the membrane lipids in Bacteria and Eukarya vs. Archaea— some scientists have proposed that the last universal common ancestor (the common ancestor to archaea and bacteria) had a heterochiral membrane (containing both mirror image forms of the glycerol phosphate backbone), and most likely both ester and ether linkages and both isoprenoid and fatty acid chains) (Wächtershäuser 2003, Lombard et al. 2012). From this ancestor, one lineage of cells arose that evolved a membrane containing just the G3P backbone— the Bacteria, while a second lineage, the Archaea, eventually developed a membrane having just the G1P backbone. Eukaryotes either evolved a bacterial-like membrane with a G3P backbone from an archaeal ancestor (that had archaeal membrane lipids with a G1P backbone) or they could have evolved from an ancestor with a heterochiral membrane. That is, it's possible that ancient archaea (or an ancestor to the archaea) still had heterochiral membranes and eukaryotes diverged from this group early on, evolving a homochiral membrane with a G3P backbone, while other archaea eventually evolved homochiral membranes with the G1P backbone (Lombard et al. 2012).
Either way, there apparently would have been a stage in this evolutionary process that involved a transition from a heterochiral membrane to a homochiral membrane. The problem is that scientists have yet to come up with a good explanation as to how this could have happened. No naturally-occurring organism on earth is known to have a heterochiral membrane (Wächtershäuser 2003). It has been proposed that heterochiral membranes are inherently unstable or otherwise unfavorable and thus unlikely to persist over evolutionary time (Wächtershäuser 2003). However, experimental studies conducted thus far have found heterochiral membranes to be both stable (Caforio et al. 2018, Fan et al. 1995, Shimada and Yamagishi 2011) and robust (Caforio et al. 2018); thus there is no known selection mechanism that would cause a heterochiral membrane to become homochiral (Villanueva et al. 2017).
Scientists have examined the genomes of the archaea that appear to be most closely related to eukaryotes, searching for clues about how the eukaryotic membrane could have evolved. Initial analyses indicate that some archaea that are apparently closely related to eukaryotes may have homochiral G3P-based membrane lipids with either ether-linked isoprenoids or mixed ether-linked isoprenoids and ester-linked fatty acids (Villanueva et al. 2017). These results are preliminary, as they are based on limited data, which were obtained from uncultured archaea (whose genomes were reconstructed from DNA segments obtained from environmental samples). However, if confirmed, the findings provide information on a potential transition stage in the evolution of the eukaryotic membrane.
Although the two domain model— the idea that eukaryotes evolved from archaea— is currently the most popular theory to describe the origin of eukaryotes, there is by no means a consensus on this issue. Many scientists would argue that the two domain model does not have enough convincing evidence behind it (at least not currently) to usurp Woese's three domains.
Perhaps the biggest issue with the two domain model is that the strongest evidence supporting it is based on genomic data from uncultured organisms. The genomes of the uncultured eukaryotic-like archaea were obtained by stitching together DNA sequences from a pool of DNA fragments extracted from a sediment sample. The overall DNA pool from which the archaeal genomes were reconstructed included DNA fragments from a multitude of microorganisms, including eukaryotes. Despite researchers best efforts to accurately reconstruct the genomes of the different organisms in the samples, it is possible (and in the opinion of some researchers, likely) that the reconstructed archaeal genomes included DNA sequences from actual eukaryotes (Da Cunha et al. 2017, Schirber 2016). Hopefully sometime in the near future scientists will develop the ability to isolate and culture the archaea of interest (i.e., to obtain genomes from pure cultures and to examine in detail the structural components and functionings of these cells), so that more definitive data can be obtained to evaluate the two domain model.
Although many recent studies have produced phylogenetic trees where eukaryotes are placed within the Archaea, the results of these analyses need to be interpreted with caution. This caution is warranted not just because the reconstructed genomes of uncultured archaeal could, by error, contain eukaryotic sequences, but because in general, determining accurate phylogenetic relationships at the base of the tree of life is considered to be extremely difficult if not impossible (Gribaldo et al. 2010, Koonin and Yutin 2014, Koonin 2015). Such analyses are notoriously prone to artifacts and biases that can affect the outcome of the trees (Koonin and Yutin 2014). The choice of proteins/genes to include in the analysis and the choice of species (e.g., whether fast evolving species are included) can determine whether the analysis results in a three domain or two domain tree (Da Cunha et al. 2017).
In conclusion, we could summarize our current state of knowledge regarding the
protoeukaryotic host cell as follows:
We know that it obtained an endosymbiont (an alphaproteobacterium), which
evolved into an organelle known as the mitochondrion. But we don't know how the
endosymbiont ended up inside the host cell. We know that the host cell
was more closely related to archaea than to bacteria, but whether the
host cell was an archaeon or whether it belonged to a separate domain—a
distinct lineage of protoeukaryotes—is still under debate.
A Theory to Explain Eukaryote Complexity
Regardless of whether the host cell was an archaeon or an early eukaryote, once it acquired a mitochondrial organelle it gained an important advantage— increased energy production—and cells with higher energy demands could simply produce more mitochondria to meet those demands. The energetic advantage provided by mitochondria is obvious when you look at cells that have high energy requirements, such as heart cells. The heart is the most energy-demanding organ in the human body and it is estimated that a 100 gram heart requires 100 times its weight in ATP per day (Dorn 2013). In order to generate sufficient ATP to power its contractions and other metabolic needs, a heart cell is packed with approximately 3,500 to 5,000 mitochondria, which occupy as much as 35% of its total cell volume (Sinatra and Houston 2015).
According to a theory proposed by Lane and Martin (2010), the evolution of the mitochondrial organelle was a crucial event in the history of life on earth— mitochondria increased the amount of energy available to cells, and this vastly increased energy budget made it possible for complex multicellular life to evolve.
Although bacteria and archaea are highly evolved organisms that have been around for a long time, they have never produced complex multicellular forms— at least no more complex than filament-forming cyanobacteria. Lane and Martin argue that this is true because these cells are incapable of supporting large genomes due to energetic limitations. Lane and Martin's hypothesis is fairly complex and is most easily explained in steps, as summarized below.
A central tenet of Lane and Martin's hypothesis is that a large genome is necessary for complex multicellular organisms to evolve. Large genomes contain noncoding DNA within and between genes, and have relatively large genes capable of encoding large proteins. A big genome provides evolution with more material to tinker with — lots of genetic material is present to allow for the evolution of regulatory elements including microRNAs, and massive numbers of new proteins may be explored and expressed in different combinations — making it possible for organisms to acquire novel complex traits. According to Lane and Martin, a large genome is one of the prerequisites "affording the cell the chance — but not the necessity— of becoming complex."[8]
Having a large genome is energetically costly. While the average cell only expends about 2 percent of its energy budget on DNA replication, it uses approximately 75 percent of its total energy budget on protein synthesis (Harold 1986). Thus cells with large genomes, which encode many proteins, require a lot more energy than similarly-sized cells with small genomes. According to Lane (2011) "there is a near linear relationship between the number of genes and the energetic cost."
-
Typically organisms with large genomes have large cells. Many studies have found a strong positive correlation between genome size and cell size, although various other factors may affect or be correlated with cell size (Mueller 2015). Changes in the amount of DNA in a cell are believed to directly affect cell volume (Kozłowski et al. 2003), and an increase in cell volume by itself requires an increased energy budget, as larger cells require more proteins (i.e., protein synthesis scales with cell volume [Lane and Martin 2010]).
Unlike eukaryotes, bacteria and archaea are unable to scale up energy production to the extent needed to support large cells with large energy-consuming genomes.
On average, bacteria and archaea have much smaller cells and much smaller genomes than eukaryotes. The volume of a eukaryotic cell often exceeds that of a bacterium or archaeon by a factor of 1,000 or more (Alberts et al. 1994) and the mass of a eukaryotic cell can be 15,000 times larger (Lane and Martin 2010). Whereas bacteria and archaea tend to have genomes of 10 megabases or less, eukaryotic genomes may easily reach sizes of 100,0000 megabases (Lane 2011), or in extreme cases over 30 gigabases (Nowoshilow et al. 2018).
Bacteria and archaea tend to have genomes that are streamlined so as to save energy. In contrast to eukaryotes, bacteria and archaea have genomes with a high gene density (due to a lack inter- and intragenic material and relatively short genes) and they have comparatively short proteins, which help to minimize energy use.
So why are bacteria and archaea apparently energetically limited to having small cell sizes and small genomes, whereas eukaryotes are not? Before answering this question, it is helpful to review some relevant background information:
- Cells rely on ATP as a source of energy.
- The vast majority of ATP is produced in membranes that contain electron transport chains and ATP synthase. The chemical reactions of the electron transport chains result in an electrochemical gradient across the membrane. Energy stored in this gradient is used to power ATP synthase— the molecular machine that produces ATP.
- In archaea and bacteria, ATP is produced in the cell membrane. In eukaryotes, ATP is not produced in the cell membrane; instead, it is synthesized in the inner membrane of mitochondria (and in the thylakoid membrane of chloroplasts in photosynthetic eukaryotes). (Membranes in which ATP is produced are referred to as "bioenergetic membranes.")
-
Eukaryotes obtain the energy needed to support large genomes from ATP production in the bioenergetic membrane of the mitochondria. Eukaryotic cells with greater energy needs can increase the surface area of bioenergetic membrane (and thus enable greater ATP production) by producing more mitochondria.
Bacteria and archaea it seems should also be able to boost ATP production by increasing the surface area of bioenergetic membrane in the cell. In this case, they would be raising ATP production by increasing the the surface area of cell membrane. According to Lane and Martin there are two main problems that result from this.
The first problem has to do with issues of scaling. An obvious way of increasing the surface area of cell membrane is to simply increase the size of the cell. However, when a cell increases in size its volume increases at a greater rate than its surface area, so for a given cell shape, the larger the cell, the smaller the surface area to volume ratio. This is a problem because the greater the volume of a cell, the more proteins the cell needs to synthesize, and the more energy it requires to make those proteins. However, at the same time the relative amount of surface area (bioenergetic membrane) where ATP can be produced shrinks. So bacteria and archaea can't just increase their cell size in order to gain enough bioenergetic membrane to support a large genome— their energy deficit will become greater as cell size increases.
So the apparent logical solution is for a cell to have a lot of infolded/internalized membrane. There are many examples of regular-sized bacteria with lots of infolded/internalized membrane, such as cyanobacteria (and the alphaproteobacterium that became the mitochondrion). To create sufficient energy to support a large genome, why not have a large cell with lots of infolded/internalized membrane?
According to Lane and Martin, regardless of whether or not a large cell has a lot of infolded/internalized membrane, there is second obstacle it has to deal with— the apparent need for core respiratory genes to be located within a short distance of bioenergetic membrane in order to maintain ATP synthesis (via oxidative phosphorylation) within the cell. More specifically, certain core genes that encode proteins of the electron transport chain need to be located near bioenergetic membrane— so that those genes can rapidly respond to, and remedy (by expressing the appropriate proteins) changes in membrane potential.
This apparent requirement is described by the CoRR hypothesis (Co-location for Redox Regulation) (Allen 2003). The CoRR hypothesis applies to both the processes of respiration and photosynthesis. Both processes involve electron transport chains, which consist of a series of reduction-oxidation (redox) reactions (when an electron is transferred from one carrier to the next in the chain, the carrier that accepts the electron becomes reduced while the carrier that donates the electron becomes oxidized). As described earlier, the chemical reactions of the electron transport chain capture energy from electrons and use that energy to create an electrochemical proton gradient across a membrane. The energy stored in the gradient is used to make ATP.
The vast majority of proteins found in mitochondria and chloroplasts are encoded by genes now found in the nucleus of the cell. However, mitochondria and chloroplasts that are capable of making ATP through use of electron transport chains always retain a core genome that includes genes that encode membrane-spanning subunits involved in electron transport and proton pumping (Allen 2003). According to the CoRR hypothesis, these genes have to stay within the organelle— they must be located near their gene products because the expression of these respiratory genes is actually controlled by their gene products (or rather by the redox state of their gene products) (Allen 2003). The core respiratory genes, their gene products, and other electron carriers with which their gene products react form part of a complex regulatory feedback loop that ensures that the energy transduction system continues to function properly.
As noted by Lane and Martin (2010), this energy transduction system needs to be under close regulatory control because failure of a mitochondrion (or chloroplast) to maintain its membrane potential (or rather electrochemical proton gradient across its membrane) could lead to catastrophic results, including plummeting ATP production, a rise in free radical leak, and programmed cell death. Thus, a mitochondrion (or chloroplast) needs to be able to respond quickly to changes in its electrochemical proton gradient and it is able to do this by retaining a core of essential genes.
So to summarize, there are two big hurdles that make it difficult for bacteria and archaea to scale up their energy budget so that they can support large cells with large genomes: (1) the fact that when a cell expands in size, its volume increases at a greater rate than its surface area. This is a problem for bacteria and archaea because they produce ATP in their cell membrane. So if they expand the surface area of cell membrane by increasing cell size, then their cell volume (and hence overall energy requirements) will increase at a greater rate than the surface area of bioenergetic membrane (and hence the rate at which ATP can be synthesized); and (2) the requirement that core respiratory genes be located close to bioenergetic membrane.
The innovation of mitochondria allowed eukaryotes to easily solve both of these problems. With the advent of mitochondria, eukaryotes no longer needed to produce ATP in their cell membranes, so they were able to escape the surface area to volume scaling problem. To make more ATP— to support larger cells with larger genomes—eukaryotes just needed to produce more mitochondria. By retaining core respiratory genes within their mitochondria, eukaryotes were able to grow large cells and yet still maintain close regulatory control over energy transduction processes.
Although they lack mitochondria, some bacteria have managed to evolve large cells containing massive amounts of DNA, thus seeming to falsify Lane and Martin's hypothesis. However, according to Lane and Martin, upon closer examination, these large bacteria are still energy-limited compared to eukaryotes, and thus they are incapable of giving rise to complex multicellular life forms.
Despite having big cells, large bacteria have small genomes—within the size range typical of other bacteria and archaea, and their genomes are similarly compacted for energy efficiency. Large bacteria contain lots of DNA not because they have large genomes, but because they have many copies of those genomes.
While eukaryotic cells normally possess a suite of complex cellular mechanisms or features, which tend to be energy-intensive (e.g., large genomes, energy-powered nutrient uptake systems, and motor proteins for moving cargo within a cell), archaea and bacteria, including large bacteria, lack these features.[9]
To achieve large cell sizes, large bacteria have had to evolve solutions to both the surface area to volume scaling problem and the need to have core respiratory genes located close to bioenergetic membrane.[10] However, these solutions are less than ideal, as they don't enable large bacteria to obtain the surplus energy needed to evolve complexity (i.e., the energy needed to grow large genomes and acquire energy-intensive cellular mechanisms).
Due to the scaling problem, many large bacteria are unable to produce enough ATP to support their full volume (i.e., a volume filled with biologically active cytoplasm). They are able to get around this problem and attain a large cell size by having a sizable portion of their cell volume occupied by vacuoles or other biologically inert components. For example, the largest bacterium currently known, Thiomargarita namibiensis, which has a diameter up to 0.8 mm, has a giant vacuole that occupies approximately 98 percent of its cell volume (Schulz and Jørgensen 2001, Mendel et al. 2008). So it requires considerably less energy (needs to make considerably fewer proteins) than an equivalently sized cell that is completely filled with [biologically active] cytoplasm.
Some large bacteria may be capable of producing comparable amounts of ATP as similarly-sized eukaryotes, by having highly infolded internalized membrane. A possible example is the cyanobacterium Synechocystis, which has extensive internal thylakoid membranes and can grow to a few micrometers in diameter (Lane 2014). However, as noted before, based on the CoRR hypothesis, all cells need to have key respiratory genes located near their bioenergetic membrane. The way this problem is handled in large bacteria is for the cell to be highly polyploid, with copies of the genome spaced out around the membrane.[11] (Large bacteria may contain hundreds, thousands, tens of thousands, and in some cases hundreds of thousands of copies of their genomes.) Although this strategy allows large bacteria to maintain regulatory control over their respiratory processes, it does so in a very inefficient and energetically costly manner compared to the method used by eukaryotes. Whereas eukaryotes utilize mitochondria, which have extremely reduced genomes (containing an estimated 0.5-1.2% of their ancestral bacterial genes [Burger et al. 2013]), large bacteria need to maintain and replicate a huge number of copies of their full genomes. Unless large bacteria and archaea can come up with a way of having just their core respiratory genes distributed around their membranes (rather than full copies of their genome), they will always be energetically limited compared to eukaryotes.
Although bacteria and archaea are incredibly efficient and successful at small cell sizes (e.g., bacteria have faster respiration rates and divide faster than eukaryotes), eukaryotes, thanks to their mitochondria, are uniquely adapted to support large cells with large genomes—and ultimately—according to Lane and Martin— complex multicellular life forms.
Groups of Eukaryotes
Some recent studies have classified eukaryotes into five (or more) supergroups (e.g., Burki 2014, Pawlowski 2013, Adl et al. 2012). These classifications are still in a state of flux—there are some taxa whose taxonomic positions are uncertain or conflicting. The so-called supergroups include the following (based on Burki 2014):
- Opisthokonta (containing animals, fungi and some heterotrophic protoctists)
- Amoebozoa (containing mostly amoeba-like heterotrophic protoctists; including mycetozoan slime molds)
- Excavata (containing heterotrophic protoctists including many anaerobes [e.g. oxymonads] and parasites [e.g. Giardia]; also euglenids [e.g., Euglena)])
- Archaeplastida (containing glaucophyte algae, red algae, and green algae and land plants)
- SAR (a clade consisting of Stramenopiles [e.g., diatoms, kelps, and water molds], Alveolates [e.g., ciliates, dinoflagellates and apicomplexans], and Rhizaria [e.g., Paulinella, radiolarians and foraminiferans])
The supergroups contain the vast majority of identified eukaryotic taxa; however there are a number of additional taxa with uncertain taxonomic status. Counting up all the groups—the supergroups plus the groups of taxa that are currently unclassifiable, yields approximately 15 or more groups of eukaryotes (Burki 2014, Pawlowski 2013, Adl et al. 2012).
For simplicity, Mass Nature divides the eukaryotes up into four groups:
- Plants (consisting of vascular plants and bryophytes);
- Animals
- Fungi (including lichens); and
- Protoctists (all of the remaining eukaryotes that don't fit into the other categories).
[1] Scientists have identified at least three different systems of cell division in archaea: one similiar to that in bacteria, which involves the protein FtsZ; one known as ESCRT-III-based (Cdv-based), a similar version of which is also involved in cell division in the Eukarya; and a third system that is possibly actin-based (Makarova et al. 2010).[Return to text]
[2] Use of 16S ribosomal RNA data has, for a long time, been considered to be a ideal way of determining evolutionary relationships among distantly related organisms. Ribosomal RNA is found in all cellular life; it performs a critical function essential to the survival of an organism: catalyzing protein synthesis, and generally has a slow evolution rate. Also, genes encoding ribosomal RNA are considered to be "informational" genes (as opposed to "operational" genes), and thus much less subject to horizontal gene transfer (Jain et al. 1999). However, recent studies have indicated that horizontal gene transfer does occasionally occur in ribosomal RNA gene segments (Kitahara and Miyazaki 2013); and, there are other problems when using ribosomal RNA, including intra-genomic heterogeneity (Rajendhran and Gunasekaran 2011), making ribosomal RNA sequence comparisons a less than perfect, although still useful means for identifying and classifying microbes.[Return to text]
[3]Although no bacteria or archaea are known to undergo phagocytosis, the bacterium Gemmata obscuriglobus (phylum Planctomycetes) has been found to be capable of uptaking protein from the environment using an energy-dependent endocytosis-like process (Lonhienne et al. 2010).[Return to text]
[4]Peptidoglycan-synthesizing MUR genes have been found in some other plants, including Arabidopsis thaliana and gymnosperms. Although the associated gene products appear to localize in the chloroplasts, the function of the MUR genes in these plants is unknown— they are not involved in chloroplast division (Lin et al. 2017).[Return to text]
[5]Of the mitochondria and MROs listed, genomes are found in aerobic, anaerobic, and hydrogen-producing mitochondria. Mitosomes and hydrogenosomes generally lack genomes (although in a few cases genomes have been found in hydrogenosomes) [Müller et al. 2012].[Return to text]
[6]Processing of the nuclear and cytosolic iron-sulfur proteins is completed by two eukaryotic systems: the mitochondrial ISC export apparatus and the cytosolic iron-sulfur protein assembly machinery (Lill and Mühlenhoff 2008).[Return to text]
[7]Of interesting note there is a representative from this group that is adapted to low oxygen environments— Stygiella incarcerata. This organism has a hydrogenosome-like organelle, which appears to lack a genome and any evidence of an aerobic energy-generating metabolism (Leger et al. 2016). The anaerobic adaptations of Stygiella's mitochondrial organelle are strikingly similar to those found in the MROs of unrelated anaerobic eukarotes. Because of these similarities, the researchers studying this organism have concluded that aerobic eukaroytes expanding into low oxygen environments may be rapidly adapting to such environments by acquiring genes from other eukaryotes via horizontal gene transfer (Leger et al. 2016). Other researchers have come to the same conclusion (e.g. Stairs et al. 2015), the reason being that the eukaryotic enzymes involved in anaerobic energy metabolism don't appear to have originated from alphaproteobacteria, i.e., no direct phylogenetic evidence has been found showing an alphaproteobacterial origin for the genes encoding these enzymes (Stairs et al. 2015). in addition, the anaerobic enzymes of eukaryotes appear to be generally missing from extant alphaproteobacteria (with the exception of taxa that appear to have recently acquired genes for the enzymes through horizontal gene transfer [Stairs et al. 2015]).[Return to text]
[8]Of course larger genomes don't necessarily produce more complex organisms — otherwise creatures such as lungfish, salamanders, and onion plants would be ruling the earth. (Although these are all rather complex organisms they are arguably less complex than humans, who have a considerably smaller [although still large] genome.) Some genomes grow quite large due to the excess accumulation of parasitic elements — genetic material that creates lots of copies of itself with little or no benefit to the host. In many organisms a lot of this seemingly useless noncoding DNA is tolerated because it doesn't appreciably affect the fitness of the host (e.g. in lungfish, salamanders, and onion plants); in other organisms — where metabolic efficiency is paramount (e.g. migrating birds)— the genome contains relatively little of this superfluous DNA.[Return to text]
[9]Occasionally a eukaryote-type feature is discovered in a bacterium or archaeon but never a suite of these energy-requiring features. Adaptations, such as complex nutrient uptake systems and motor proteins may not be all that useful in normal (small)-sized bacteria and archaea— because in small-sized cells, molecular diffusion (which requires no additional energy input) is a very efficient and effective means for obtaining substrates from the environment and for moving substances to desired locations within a cell. However, at larger cell sizes it becomes much more difficult (takes much longer) for cells to rely on molecular diffusion alone for these needs.[Return to text]
[10]There are other major problems that large bacteria and archaea have to deal with: because they rely on molecular diffusion for nutrient uptake and transport within the cell, they are limited by the rate of diffusion of substrates across their cell membrane (larger cells need more substrates but there is proportionately less surface area through which those substrates can diffuse) and a lot more time is required for molecules and substrates to move to their needed locations in large-sized cells (Schulz and Jørgensen 2001). Eukaryotes don't have these cell size constraints, in part because they have evolved sophisticated nutrient uptake systems and they are able to actively transport substances within the cell through use of motor proteins on cytoskeletal filaments (Mendel et al. 2008). However these eukaryotic solutions come at a cost— they require extra energy, which may be the reason why such useful innovations are not normally found in bacteria and archaea.[Return to text]
[11]Another reason why large bacteria would need to have copies of their genome spaced out around the membrane is that they don't have an active means of moving substances within their cells (i.e., they rely on molecular diffusion, which becomes ineffective at large cell sizes). Large bacteria need to synthesize many membrane proteins. If the cell's genetic material and protein synthesis machinery are centrally located, there is no good way to move the proteins to the locations needed. So why don't bacteria have motor proteins like eukaryotes? A possible reason is that motor proteins require extra ATP and bacteria don't have a surplus of energy to support them.[Return to text]
Sources
Adl, S.M., A.G. Simpson, C.E. Lanec, J. Lukeš, D. Basse, S.S. Bowser, M. Brown, F. Burki, M. Dunthorn, V. Hampl, A. Heiss, M. Hoppenrath, E. Lara, L. leGall, D.H. Lynn, H. McManus, E.A.D. Mitchell, S.E. Mozley-Stanridge, L. Wegener Parfrey, J. Pawlowski, S. Rueckert, L. Shadwick, C. Schoch, A. Smirnov, and F.W. Spiegel. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012 September; 59(5):429–493. doi:10.1111/j.1550-7408.2012.00644.x.
Alberts, B., D. Bray, J. Lewis, M. Raff, K. Roberts, and J.D. Watson. 1994. Molecular Biology of the Cell. Third Edition. Garland Publishing, Inc. New York. 1294 pp.
Allen, J.F. 2003. The function of genomes in bioenergetic organelles. Phil. Trans. R. Soc. Lond. B. 358:19-38.
Aminov, R.I. 2013. Role of archaea in human disease. Front Cell Infect Microbiol. 2013 Aug 13;3:42. doi: 10.3389/fcimb.2013.00042. eCollection 2013.
Archibald, J.M. 2015. Endosymbiosis and Eukaryotic Cell Evolution. Current Biology. 25:R911-R921.
Arimura, S., and N. Tsutsumi. 2002. A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. PNAS. 99(8):5727-5731.
Bapteste, E., M.A. O'Malley, R.G. Beiko, M. Ereshefsky, J.P. Gogarten, L. Franklin-Hall, F. Lapointe, J. Dupré, T. Dagan, Y. Boucher, and W. Martin. 2009. Prokaryotic evolution and the tree of life are two different things. Biology Direct. 4:34. doi:10.1186/1745-6150-4-34.
Bapteste, E., L. van Iersel, A. Janke, S. Kelchner, S. Kelk, J.O. McInerney, D.A. Morrison, L. Nakhleh, M. Steel, L. Stougie, and J. Whitfield. 2013. Networks: expanding evolutionary thinking. Trends in Genetics. 29(8):439-441. August 2013.
Barillà, D. 2016. Driving Apart and Segregating Genomes in Archaea. Trends Microbiol. 24(12):957-967.
Beech, P.L., T. Nheu, T. Schultz, S. Herbert, T. Lithgow, P. Gilson, and G.I. McFadden. 2000. Mitochondrial FtsZ in a Chromophyte Alga. Science. 287:1276-1279.
Beisson, J. 2008. Preformed cell structure and cell heredity. Prion. 2(1):1-8.
Berenguer, J., F. Rojo, M.A. de Pedro, B. Pfanzagl, and W. Löffelhardt. 1987. Penicillin-binding proteins in the cyanelles of Cyanophora paradoxa, a eukaryotic photoautotroph sensitive to b-lactam antibiotics. FEBS Lett. 224:401–405.
Blaustein, R. 2016. The Great Oxidation Event. Evolving understandings of how oxygenic life on Earth began. BioScience. 66(3):189-195.
Bonen, L. and W.F. Doolittle. 1975. On the Prokaryotic Nature of Red Algal Chloroplasts. Proc. Nat. Acad. Sci. USA. 72(6):2310-2314.
Boto, L. 2010. Horizontal gene transfer in evolution: facts and challenges. Proceedings of the Royal Society B. 277:819-827. doi:10.1098/rspb.2009.1679.
Böttger, E.C., B. Springer, T. Prammananan, Y. Kidan and P. Sander. 2001. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Reports. 2(4):318-323.
Boxma, B., R.M. de Graaf, G.W.M. van der Staay, T.A. van Alen, G. Ricard, T. Gabaldón, A.H.A.M. van Hoek, S.Y. Moon-van der Staay, W.J.H. Koopman, J.J. van Hellemond, A.G.M. Tielens, T. Friedrich, M. Veenhuis, M.A. Huynen, and J.H.P. Hackstein. 2005. An anaerobic mitochondrion that produces hydrogen. Nature. 434(7029):74–79.
Buick, R. 2008. When did oxygenic photosynthesis evolve? Philosophical Transactions of the Royal Society B. 363(1504):2731-2743. doi: 10.1098/rstb.2008.0041.
Burger, G., M.W. Gray, L. Forget, and B.F. Lang. 2013. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 5(2):418-438.
Burki, F. 2014. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol. 2014;6:a016147.
Burki, F., B. Imanian, E. Hehenberger, Y. Hirakawa, S. Maruyama, and P.J. Keeling. 2014. Endosymbiotic gene transfer in tertiary plastid-containing dinoflagellates. Eukaryotic Cell. 13(2):246-255.
Caforio, A., M.F. Siliakus, M. Exterkate, S. Jain, V.R. Jumde, R.L.H. Andringa, S.W.M. Kengen, A.J. Minnaard, A.J.M. Driessen, and J. van der Oost. 2018. Converting Escherichia coli into an archaebacterium with a hybrid heterochiral membrane. Proc Natl Acad Sci USA. 115(14):3704-3709.
Cavalier-Smith, T. 2006. Origin of mitochondria by intracellular enslavement of a photosynthetic purple bacterium. Proceedings of the Royal Society B. 273:1943-1952.
Celler, K., R.I. Koning, A. J. Koster, and G.P. van Wezel. 2013. Multidimensional View of the Bacterial Cytoskeleton. Journal of Bacteriology. 195(8):1627-1636.
Chaban, B., S.Y.M Ng, and K.F. Jarrell. 2006. Archaeal habitats - From the extreme to the ordinary. Canadian journal of microbiology. 52: 73-116. 10.1139/w05-147.
CPN (Committee on Phylogenetic Nomenclature) 2010. International Code of Phylogenetic Nomenclature. Version 4c. Most recent revision: January 12, 2010. Available from: https://www.ohio.edu/phylocode/index.html. Accessed September 29, 2017.
Cox, C.J., P.G. Foster, R.P. Hirt, S.R. Harris, and T.M. Embley. 2008. The archaebacterial origin of eukaryotes. Proc. Natl Acad. Sci. USA. 105(51):20356-20361.
Da Cunha V., M. Gaia, D. Gadelle, A. Nasir, and P. Forterre. 2017. Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet 13(6):e1006810. https://doi.org/10.1371/journal.pgen.1006810.
Davidov, Y., D. Huchon, S.F. Koval, and E. Jurkevitch. 2006. A new α-proteobacterial clade of Bdellovibrio-like predators: implications for the mitochondrial endosymbiotic theory. Environ Microbiol. 8(12):2179-2188.
Delaye, L., C. Valadez-Cano, and B. Pérez-Zamorano. 2016. How Really Ancient Is Paulinella Chromatophora?. PLOS Currents Tree of Life. 2016 Mar 15 . Edition 1. doi: 10.1371/currents.tol.e68a099364bb1a1e129a17b4e06b0c6b.
Delwiche, C.F. and J.D. Palmer 1996. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol Biol. Evol. 13(6):873-882.
Dismukes, G.C., V.V. Klimov, S.V. Baranov, Y.N. Kozlov, J. DasGupta, and A. Tyryshkin. 2001. The origin of atmospheric oxygen on Earth: The innovation of oxygenic photosynthesis. PNAS. 98(5):2170-2175.
Doolittle, W.F. 1999. Phylogenetic classification and the universal tree. Science. 284:2124-2128.
Dorn, G.W. II. 2013. Mitochondrial dynamics in heart disease. Biochim Biophys Acta. 1833(1):233-241.
Douglas, S., S. Zauner, M. Fraunholz, M. Beaton, S. Penny, L. Deng, X. Wu, M. Reith, T. Cavalier-Smith, and U. Maier. 2001. The highly reduced genome of an enslaved algal nucleus. Nature. 410:1091-1096.
du Lac, M., A.H. Scarpelli, A.K.D. Younger, D.G. Bates, and J.N. Leonard. 2017. Predicting the dynamics and heterogeneity of genomic DNA content within bacterial populations across variable growth rates. ACS Synthetic Biology. 6:1131-1139.
Egan, E.S., M.A. Fogel, and M.K. Waldor 2005. Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes. Molecular Microbiology. 56(5):1129-1138.
Ettema, T.J.G., A.-C. Linda˚s, and R. Bernander. 2011. An actin-based cytoskeleton in archaea. Mol. Microbiol. 80:1052–1061.
Fan, Q., A. Relini, D. Cassinadri, A. Gambacorta, and A. Gliozzi. 1995. Stability against temperature and external agents of vesicles composed of archael bolaform lipids and egg PC. Biochim Biophys Acta. 1240(1):83-88.
Fenchel, T. and B.J. Finlay. 1995. Ecology and Evolution in Anoxic Worlds. Oxford University Press: New York. 276 pages.
Feniouk, B.A. 2012. ATP synthase FAQ. Updated 04 July 2012. Available from http://www.atpsynthase.info/FAQ.html.
Foster, P.G., C.J. Cox, and T.M. Embley. 2009. The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods. Philos Trans R Soc Lond B Biol Sci. 364(1527):2197-2207.
Friedrich, C.G., D. Rother, F. Bardischewsky, A. Quentmeier, and J. Fischer. 2001. Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. environ. Microbiol. 67(7):2873-2882.
Gattinger, A., M. Schloter, and J.C. Munch. 2002. Phospholipid etherlipid and phospholipid fatty acid fingerprints in selected euryarchaeotal monocultures for taxonomic profiling. FEMS Microbiol Lett. 213(1):133-139.
Gilson, P.R., U. Maier, and G.I. McFadden. 1997. Size isn't everything: lessons in genetic miniaturisation from nucleomorphs. Current Opinion in Genetics & Development. 7:800-806.
Gouy, R., D. Baurain, and H. Philippe. 2015. Rooting the tree of life: the phylogenetic jury is still out. Phil. Trans. R. Soc. B. 370:20140329. http://dx.doi.org/10.1098/rstb.2014.0329.
Gralnick, J.A. and D.K. Newman. 2007. Extracellular respiration. Mol. Microbiol. 65(1):1-11.
Gray, M.W. 2001. The origin and early evolution of mitochondria. Genome Biology, 2(6):reviews 1018.1-1018.5.
Gray, M.W. 2012. Mitochondrial Evolution. Cold Spring Harb Perspect Biol. 2012;4:a011403.
Gray, M.W. 2015. Mosaic nature of the mitochondrial proteome: implications for the origin and evolution of mitochondria. PNAS, 112(33):10133-10138. https://doi.org/10.1073/pnas.1421379112.
Gribaldo, S., A.M. Poole, V. Daubin, P. Forterre, and C. Brochier-Armanet. 2010. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nature Rev. Microbiol. 8:743-752.
Grüber,G., M.S.S. Manimekalai, F. Mayer, and V. Müller. 2014. ATP synthases from archaea: the beauty of a molecular motor. Biochimica et Biophysica Acta. 1837:940-952.
Guerrero, R., C. Pedrós-Alió, I. Esteve, J. Mas, D. Chase, and L. Margulis. 1986. Predatory prokaryotes: predation and primary consumption evolved in bacteria. Proc. Natl. Acad. Sci. USA. 83:2138-2142.
Harold, F.M. 1986. The Vital Force: A Study of Bioenergetics. Freeman, New York.
Harold, F.M. 2010. On the continuity of biological membranes. Available from: http://schaechter.asmblog.org/schaechter/2010/03/on-the-continuity-of-biological-membranes.html.
Harris, J.K., S.T. Kelley, G.B. Spiegelman, and N.R. Pace. 2003. The Genetic Core of the Universal Ancestor. Genome Research. 13(3):407–412. http://doi.org/10.1101/gr.652803
Hartman, H. and A. Fedorov. 2002. The origin of the eukaryotic cell:a genomic investigation. Proc. Natl Acad. Sci. USA. 99:1420–1425.
Heiss, A.A. and P.J. Keeling. 2006. The phylogenetic position of the oxymonad Saccinobaculus based on SSU rRNA. Protist. 157(3):335-344.
Hirano, T., K. Tanidokoro, Y. Shimizu, Y. Kawarabayasi, T. Ohshima, M. Sato, S. Tadano, H. Ishikawa, S. Takio, K. Takechi, and H. Takano. 2016. Moss chloroplasts are surrounded by a peptidoglycan wall containing D-amino acids. The Plant Cell. 28:1521-1532.
Hong, S., K.A. Harris, K.D. Fanning, K.L. Sarachan, K.M. Frohlich, and P. Agris. 2015. Evidence that antibiotics bind to human mitochondrial ribosomal RNA has amplications for aminoglycoside toxicity. The Journal of Biological Chemistry. 290(31):19273-10286.
Howe, C.J., A.C. Barbrook, R.E.R. Nisbet, P.J. Lockhart, and A.W.D. Larkum. 2008. The origin of plastids. Phil. Trans. R. Soc. B. 363:2675-2685.
Huang, C.Y., M.A. Ayliffe, and J.N. Timmis. 2003. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature. 422:72-76.
Husnik, F. and J.P. McCutcheon. 2016. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. PNAS. 113(37):E5416-E5424. doi:10.1073/pnas.1603910113.
Imanian, B. and P.J. Keeling. 2007. The dinoflagellates Durinskia baltica and Kryptoperidinium foliaceum retain functionally overlapping mitochondria from two evolutionarily distinct lineages. BMC Evolutionary Biology. 7:172. https://doi.org/10.1186/1471-2148-7-172.
Jackson C., S. Clayden, and A. Reyes-Prieto. 2015. The glaucophyta: the blue-green plants in a nutshell. Acta Societatis Botanicorum Poloniae. 84(2):149-165.
Jain, R., M.C. Rivera and J.A. Lake. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci USA. 96(7):3801-3806.
Janouškovec, J., S-L Liu, P.T. Martone, W. Carré, C. Leblanc, J. Collén, and P.J. Keeling. 2013. Evolution of Red Algal Plastid Genomes: Ancient Architectures, Introns, Horizontal Gene Transfer, and Taxonomic Utility of Plastid Markers. PLoS one: 2013; 8(3): e59001.
Jékely, G. 2014. Origin and evolution of the self-organizing cytoskeleton in the network of eukaryotic organelles. Cold Spring Harb Perspect Biol. 2014;6:a016030. doi: 10.1101/cshperspect.a016030.
Jonckheere, A.I., J.A.M. Smeitink, and R.J.T. Rodenburg. 2012. Mitochondrial ATP synthase: architecture, function and pathology. J. Inherit Metab Dis. 35(2):211-225.
Kalghatgi, S., C.S. Spina, J.C. Costello, M. Liesa, J.R. Morones-Ramirez, S. Slomovic, A. Molina, O.S. Shirihai, and J.J. Collins. 2013. Bactercidal antibiotics induce mitochondrial dysfunction and oxidative damage in mammalian cells. Science Translational Medicine, 5(192), 192ra85. http://doi.org/10.1126/scitranslmed.3006055.
Karnkowska, A., and V. Hampl. 2016. The curious case of vanishing mitochondria. Microbial Cell. 3(10):491-494. doi: 10.15698/mic2016.10.531.
Karnkowska, A., V. Vacek, Z. Zubáčová, S.C. Treitli, R. Petrželková, L. Eme, L. Novák, V. Žárský, L.D. Barlow, E.K. Herman, P. Soukal, M. Hroudová, P. Doležal, C.W. Stairs, A.J. Roger, M. Eliáš, J.B. Dacks, Č. Vlček, and V. Hampl. 2016. A Eukaryote without a Mitochondrial Organelle. Current Biology. 26:1274-1284.
Katayama, N., H. Takano, M. Sugiyama, S. Takio, A. Sakai, K. Tanaka, H. Kuroiwa, and K. Ono. 2003. Effects of antibiotics that inhibit the bacterial peptidoglycan synthesis pathway on moss chloroplast division. Plant Cell Physiol. 44:776–781.
Kaut, A., H. Lange, K. Diekert, G. Kispal and R. Lill. 2000. Isa1p is a component of the mitochondrial machinery for maturation of cellular iron-sulfur proteins and requires conserved cysteine residues for function. The Journal of Biological Chemistry. 275:15955-15961. doi: 10.1074/jbc.M909502199.
Keeling, P.J. 2004. Diversity and evolutionary history of plastids and their hosts. American Journal of Botany. 91(10):1481-1493.
Keeling, P.J. 2010. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 365(1541):729-748.
Khelaifia, S. and M. Drancourt. 2012. Susceptibility of archaea to antimicrobial agents: applications to clinical microbiology. Clinical Microbiology and Infection. 18:841-848.
Kitahara, K. and K. Miyazaki. 2013. Revisiting bacterial phylogeny, Mobile Genetic Elements, 3:1, e24210, DOI: 10.4161/mge.24210
Koga Y. and H. Morii. 2007. Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol Mol Biol Rev. 71:97–120.
Koga, Y., N. Sone, S. Noquchi, and H. Morii. 2003. Transfer of pro-R hydrogen from NADH to dihydroxyacetonephosphate by sn-glycerol-1-phosphate dehydrogenase from the archaeon methanothermobacter thermautotrophicus. Biosci Biotechnol Biochem. 67(7):1605-1608.
Koonin, E.V. 2015. Origin of eukaryotes from within archaea, archaeal eukaryome and bursts of gene gain: eukaryogenesis just made easier? Phil. Trans. R. Soc. B. 370:20140333. http://dx.doi.org/10.1098/rstb.2014.0333.
Koonin, E.V. and N. Yutin. 2014. The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harb Perspect Biol. 6(4):a016188. doi:10.1101/cshperspect.a016188.
Kozłowski, J., M. Konarzewski, and A.T. Gawelczyk. 2003. Cell size as a link between noncoding DNA and metabolic rate scaling. PNAS. 100(24):14080-14085.
Kunin, V., L. Goldovsky, N. Darzentas, and C.A. Ouzounis. 2005. The net of life: reconstructing the microbial phylogenetic network. Genome Research. 15(7):954-959.
Kurland C.G. and S.G. Andersson. 2000. Origin and evolution of the mitochondrial proteome. Microbiol Mol Biol Rev. 64(4):786–820. [PubMed: 11104819]
Kuzminov, A. 2013. The chromosome cycle of prokaryotes. Mol. Microbiol. 90(2): 214-227. doi: 10.1111/mmi/12372.
Lake, J.A. 1988. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature. 331:184–186.
Lane, N. 2011. Energetics and genetics across the prokaryote-eukaryote divide. Biology Direct. 6:35. http://www.biology-direct.com/content/6/1/35.
Lane, N. 2014. Bioenergetic constraints on the evolution of complex life. Cold Spring Harb Perspect Biol. 2014;6a015982.
Lane, N. 2015. The Vital Question. Energy, Evolution, and the Origins of Complex Life. W.W. Norton & Company. New York. 368 pages.
Lane, N. and W. Martin. 2010. The energetics of genome complexity. Nature 467:929-934.
Lang, B.F., G. Burger, C.J. O'Kelly, R. Cedergren, G.B. Golding, C. Lemieux, D. Sankoff, M. Turmel, and M.W. Gray. 1997. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387:493-497.
Leger, M.M., L. Eme, L.A. Hug, and A.J. Roger. 2016. Novel hydrogenosomes in the microaerophilic jakobid Stygiella incarcerata. Mol Biol Evol. 33(9):2318-2336. doi: 10.1093/molbev/msw103.
Lill, R. and U. Mühlenhoff. 2008. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77:669-700.
Lin, X., N. Li, H. Kudo, Z. Zhang, J. Li, L. Wang, W. Zhang, K. Takechi, and H. Takano. 2017. Genes Sufficient for Synthesizing Peptidoglycan are Retained in Gymnosperm Genomes, and MurE from Larix gmelinii can Rescue the Albino Phenotype of Arabidopsis MurE Mutation. Plant Cell Physiol. 58(3):587-597.
Lindås, A.-C., E.A. Karlsson, M.T. Lindgren, T.J. Ettema, and R. Bernander. 2008. A unique cell division machinery in the Archaea. Proc. Natl Acad. Sci. USA. 105:18942– 18946.
Lindsay, M.R., R.I. Webb, M. Strous, M.S. Jetten, M.K. Butler, R.J. Forde, and J.A. Fuerst. 2001. Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch. Microbiol. 175(6):413–429.
Lombard, J., P. López-García, and D. Moreira. 2012. The early evolution of lipid membranes and the three domains of life. Nat Rev Microbiol. 10:507–515.
Lonhienne, T.G.A., E. Sagulenko, R.I. Webb, K. Lee, J. Franke, D.P. Devos, A. Nouwens, B.J. Carroll, and J.A. Fuerst. 2010. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. PNAS, 107(29):12883-12888.
Ludwig, M. and S.P. Gibbs. 1989. Evidence that the nucleomorphs of Chlorarachnion reptans (Chlorarachniophyceae) are vestigial nuclei: morphology, division and DNA-DAPI fluorescence. Journal of Phycology, 25(2):385-394.
Machida, M., K. Takechi, H. Sato, S.J. Chung, H. Kuroiwa, S. Takio, M. Seki, K. Shinozaki, T. Fujita, M. Hasebe, and H. Takano. 2006. Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss. PNAS. 103(17):6753-6758.
Maguire, F. and T.A. Richards. 2014. Organelle evolution: a mosaic of 'mitochondrial' functions. Current Biology. 24(11):R518-R520.
Makarova, K.S., N. Yutin, S.D. Bell, and E.V. Koonin. 2010. Evolution of diverse cell division and vesicle formation systems in Archaea. Nature Reviews. Microbiology. 8(10):731–741. http://doi.org/10.1038/nrmicro2406
Maldonado, R., J. Jiménez, and J. Casadesús. 1994. Changes of ploidy during the Azotobacter vinelandii growth cycle. Journal of Bacteriology. 176(13):3911-3919.
Manson, M.D., P. Tedesco, H.C. Berg, F.M. Harold, and C. van der Drift. 1977. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci USA. 74(7):3060-3064.
Margolin, W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 6(11):862-871.
Marin, B., E.C.M. Nowack, and M. Melkonian. 2005. A plastid in the making: evidence for a second primary endosymbiosis. Protist. 156:425-432.
Martin, W. 2003. Gene transfer from organelles to the nucleus: frequent and in big chunks. PNAS. 100(15):8612-8614.
Martin, W. 2005. The missing link between hydrogenosomes and mitochondria. Trends Microbiol. 13:457–459.
Martin, W.F., S. Garg, and V. Zimorski. 2015. Endosymbiotic theories for eukaryote origin. Phil. Trans. R. Soc. B. 370:20140330.
Martin, W., T. Rujan, E. Richly, A. Hansen, S. Cornelsen, T. Lins, D. Leister, B. Stoebe, M. Hasegawa, and D. Penny. 2002. Evolutionary analysis of Arabidopsis, cyanobacterial and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. PNAS. 99(19):12246-12251.
Mavárez, J., C.A. Salazar, E. Bermingham, C. Salcedo, C.D. Jiggins and M. Linares. 2006. Speciation by hybridization in Heliconius butterflies. Nature. 441:868-871.
Mendell, J.E., K.D. Clements, J. H. Choat, and E.R. Angert. 2008. Extreme polyploidy in a large bacterium. PNAS. 105(18):6730-6734.
Miyagishima, S. 2011. Mechanism of plastid division: from a bacterium to an organelle. Plant Physiology. 155:1533-1544.
Miyagishima, S., Y. Kabeya, C. Sugita, M. Sugita, and T. Fujiwara. 2014. DipM is required for peptidoglycan hydrolysis during chloroplast division. BMC Plant Biology. 14:57.
Moreira, D., H. Le Guyader, and H. Philippe. 2000. The origin of red algae and the evolution of chloroplasts. Nature. 405:69-72.
Moseley, J.B. 2013. An expanded view of the eukaryotic cytoskeleton. Molecular Biology of the Cell. 24:1615-1618.
Mueller, R.L. 2015. Genome biology and the evolution of cell-size diversity. Cold Spring Harb Perspect Biol. 2015;7:a019125.
Müller, M., M. Mentel, J.J. van Hellemond, K. Henze, C. Woehle, S.B. Gould, R. Yu, M. van der Glezen, A.G.N. Tielens, and W.F. Martin. 2012. Biochemistry and Evolution of Anaerobic Energy Metabolism in Eukaryotes. Microbiology and Molecular Biology Reviews. 76(2):444-495.
Muñoz-Gómez, S.A., C.H. Slamovits, J.B. Dacks, K.A. Baier, K.D. Spencer, and J.G. Wideman. 2015. Ancient Homology of the Mitochondrial Contact Site and Cristae Organizing System Points to an Endosymbiotic Origin of Mitochondrial Cristae. Current Biology. 25:1489-1495.
Muñoz-Gómez, S.A., J.G. Wideman, A.J. Roger, and C.H. Slamovits. 2017. The origin of mitochondrial cristae from alphaproteobacteria. Mol Biol Evol. 34(4):943-956.
Niederman, R. 2006. Structure, Function and Formation of Bacterial Intracytoplasmic Membranes. Complex Intracellular Structures in Prokaryotes. Microbiology Monographs. 2. 193-227. 10.1007/7171_025.
Nilsson, A. and J. Nielsen. 2016. Metabolic trade-offs in yeast are caused by F1F0-ATP synthase. Scientific Reports. 6:22264. DOI: 10.1038/srep22264. Available from https://www.nature.com/articles/srep22264.pdf.
Nkamga, V.D., B. Henrissat, and M. Drancourt. 2017. Archaea: Essential inhabitants of the human digestive microbiota. Human Microbiome Journal. 3:1-8. March 2017. Available from: http://www.sciencedirect.com/science/article/pii/S2452231716300148.
Nowack, E.C., M. Melkonian, and G. Glöckner. 2008. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr Biol. 18:410–418.
Nowack, E.C.M., H. Vogel, M. Groth, A.R. Grossman, M. Melkonian, and G. Glöckner. 2011. Endosymbiotic Gene Transfer and Transcriptional Regulation of Transferred Genes in Paulinella chromatophora. 2011. Molecular Biology and Evolution. 28(1):407-422.
Nowoshilow, S., S. Schloissnig, J. Fei, A. Dahl, A.W.C. Pang, M. Pippel, S. Winkler, A.R. Hastie, G. Young, J.G. Roscito, F. Falcon, D. Knapp, S. Powell, A. Cruz, H. Cao, B. Habermann, M. Miller, E.M. Tanaka, and E.W. Myers. 2018. The axolotl genome and the evolution of key tissue formation regulators. Nature. 554:50-55.
Pawlowski, J. 2013. The new micro-kingdoms of eukaryotes. BMC Biology. 11:40. https://doi.org/10.1186/1741-7007-11-40.
Pecoraro, V., K. Zerulla, C. Lange, and J. Soppa. 2011. Quantification of Ploidy in Proteobacteria Revealed the Existence of Monoploid, (Mero-)Oligoploid and Polyploid Species. PLoS ONE. 6(1): e16392.
Pfeiffer, T. and A. Morley. 2014. An evolutionary perspective on the Crabtree effect. Frontiers in Molecular Biosciences. Vol. 1, Article 17. doi: 10.3389/fmolb.2014.00017.
Pfeiffer, T., S. Schuster, and S. Bonhoeffer. 2001. Cooperation and competition in the evolution of ATP-producing pathways. Science. 292:504-507.
Philippe, H., H. Brinkmann, D.V. Lavrov, D.T.J. Littlewood, M. Manuel, G. Wörheide, and D. Baurain. 2011. Resolving Difficult Phylogenetic Questions: Why More Sequences Are Not Enough. PLoS Biol. 9(3):e1000602. https://doi.org/10.1371/journal.pbio.1000602
Pirbadian, S., S.E. Barchinger, K.M. Leung, H.S. Byun, Y. Jangir, R.A. Bouhenni, S.B. Reed, M.F. Romine, D.A. Saffarini, L. Shi, Y.A. Gorby, J.H. Golbeck, and M.Y. El-Naggar. 2014. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. PNAS. 111(35):12883-12888. https://doi.org/10.1073/pnas.1410551111.
Pogozheva, I.D., S.Tristram-Nagle, H.I Mosberg, and A.L. Lomize. 2013. Structural adaptations of proteins to different biological membranes. Biochim Biophys Acta. 1828(11):2592-2608.
Portail, M., C. Brandily, C. Cathalot, A. Colaço, Y. Gélinas, B. Husson, P. Sarradin, and J. Sarr. 2018. Food-web complexity across hydrothermal vents on the Azores triple junction. Deep Sea Research Part I: Oceanographic Research Papers. 131:101-120.
Prescott, L.M., J.P. Harley, and D.A. Klein. 1993. Microbiology. Second Edition. Volume one. Wm. C. Brown Publishers, Dubuque, Iowa. 126 pages.
Probst, A.J., A.K. Auerbach, and C. Moissl-Eichinger. 2013. Archaea on Human Skin. PLoS One. 8(6): e65388. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3680501/.
Rajendhran, J. and P. Gunasekaran. 2011. Microbial phylogeny and diversity: Small subunit ribosomal RNA sequence analysis and beyond. Microbiological Research. 166(2):99-110.
Raven, J.A. and J.F. Allen. 2003. Genomics and chloroplast evolution: what did cyanobacteria do for plants? Genome Biology. 4(3):209
Raven, P.H., R.F. Evert, and S.E. Eichhorn. 1992. Biology of Plants. Fifth edition. Worth Publishers: New York. 791 pages.
Rivera M.C., R. Jain, J.E. Moore, and J.A. Lake. 1998. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci USA. 95(11):6239-6244.
Rogers, M.B., P.R. Gilson, V. Su, G.I. McFadden, and P.J. Keeling. 2007. The complete chloroplast genome of the Chlorarachniophyte Bigelowiella natans: evidence for independent origins of Chlorarachniophyte and Euglenid secondary endosymbionts. Molecular Biology and Evolution. 24(1):54-62.
Rothschild, L.J. 2008. The evolution of photosynthesis...again? Phil. Trans. R. Soc. B. 363:2787-2801.
Rouhan, G. and M. Gaudeul. 2014. Plant Taxonomy: A Historical Perspective, Current Challenges, and Perspectives. In Methods in Molecular Biology (Clifton, N.J.). January 2014. Volume 1115: 1-37.
Sagulenko E., A. Nouwens, R.I. Webb, K. Green, B. Yee, G. Morgan, A. Leis, K.-C. Lee, M.K. Butler, N. Chia, U.T. Phuong Pham, S. Lindgreen, R. Catchpole, A.M. Poole, and J.A. Fuerst. 2017. Nuclear Pore-Like Structures in a Compartmentalized Bacterium. PLoS ONE. 12(2):e0169432. https://doi.org/10.1371/journal.pone.0169432
Sapp, J. 2005. The Prokaryote-Eukaryote Dichotomy: Meanings and Mythology. Microbiology and Molecular Biology Reviews. 69(2):292-305.
Schirber, M. 2016. A disputed origin for eukaryotes. NASA Astrobiology Newsletter. March 14, 2016. Available from: https://astrobiology.nasa.gov/news/a-disputed-origin-for-eukaryotes/.
Schulz, H.N. and B.B. Jørgensen. 2001. Big bacteria. Annu. Rev. Microbiol. 55:105-137.
Seitz, K.W., C.S. Lazar, K.U. Hinrichs, A.P. Teske, and B.J. Baker. 2016. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J. 10(7):1696-1705.
Shi, L., T.C. Squier, J.M. Zachara, and J.K. Fredrickson. 2007. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Molecular Microbiology. 65(1):12-20.
Shiflett, A. and P.J. Johnson. 2010. Mitochondrion-related organelles in parasitic eukaryotes. Annu Rev Microbiol. 64:409-429. doi:10.1146/annurev.micro.62.081307.162826.
Shih, P.M. and N. Matzke. 2013. Primary endosymbiosis events date to the later Proterozoic with cross-calibrated phylogenetic dating of duplicated ATPase proteins. PNAS. 110(30):12355-12360.
Shimada, H. and A. Yamagishi. 2011. Stability of heterochiral hybrid membrane made of bacterial sn-G3P lipids and archaeal sn-G1P lipids. Biochemistry. 50(19):4114–4120.
Siliakus, M.F., J. van der Oost, and S.W.M. Kengen. 2017. Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles. 21(4):651-670.
Sinatra, S.T. and M. Houston (eds.). 2015. Nutritional and Integrative Strategies in Cardiovascular Medicine. CRC Press. Boca Raton, FL.
Sojo, V., A. Pomiankowski, and N. Lane. 2014. A Bioenergetic Basis for Membrane Divergence in Archaea and Bacteria. PLOS Biology. 12(8):1-12. e1001926.
Spang, A., J.H. Saw, S.L. Jørgensen, K. Zaremba-Niedzwiedzka, J. Martijn, A.E. Lind, R. van Eijk, C. Schleper, L. Guy, and T.J.G. Ettema. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 521:173-179.
Speijer, D., J. Lukeš, and M. Eliáš. 2015. Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. PNAS. 112(29):8827-8834.
Spribille, T., V. Vuovinen, P. Resl, D. Vanderpool, H. Wolinski, M. C. Aime, K. Schneider, E. Stabentheiner, J. Toome-Heller, G. Thor, Helmut Mayrhofer, H. Johannesson, and J.P. McCutcheon. 2016. Basidiomycete yeasts in the cortex of ascomycete macrolichens. Science. 353(6298): 488-492.
Stairs, C.W., M.M. Leger and A.J. Roger. 2015. Diversity and origins of anaerobic metabolism in mitochondria and related organelles. Phil. Trans. R. Soc. B. 370:20140326. http://dx.doi.org/10.1098/rstb.2014.0326.
Stoebe, B. and K.V. Kowallik. 1999. Gene-cluster analysis in chloroplast genomics. Trends in Genetics. 15(9):344-347.
Striepen, B., M. Crawford, M. Shaw, L.D. Tilney, F. Seeber, and D.S. Roos. 2000. The plastid of Toxoplasma gondii is divided by association with the centrosomes J. Cell Biol. 151:1423-1434
Szklarczyk, R. and M. Huynen. 2010. Mosaic origin of the mitochondrial proteome. Proteomics. 10:4012–4024.
Thorsness, P.E. and T.D. Fox. 1990. Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature. 346:376-379.
Timmis, J.N., M.A. Ayliffe, C.Y. Huang, and W. Martin. 2004. Endosymbiotic gene transfer:Organelle genomes forge eukaryotic chromosomes. Nature Reviews Genetics. 5(2):123–135.
Tsaousis, A.D., E.R. Kunji, A.V. Goldberg, J.M. Lucocq, R.P. Hirt, and T.M. Embley. 2008. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature. 453:553–556.
van Hoek, M.J.A. and R.M.H. Merks. 2012. Redox balance is key to explaining full vs. partial switching to low-yield metabolism. BMC Systems Biology. 6:22. http://www.biomedcentral.com/1752-0509/6/22.
Verhees, C.H., S.W.M. Kengen, J.E. Tuininga, G.J. Schut, M.W.W. Adams, W.M. DeVos, and J. Van der Osst. 2003. The unique features of glycolytic pathways in Archaea. Biochem. J. 375:231-246.
Villanueva, L., S. Schouten, and J.S. Damsté. 2017. Phylogenomic analysis of lipid biosynthetic genes of Archaea shed light on the 'lipid divide'. Environ. Microbiol. 19(1):54-69. doi: 10.1111/1462-2920.13361. Epub 2016 Jul 7.
Vitha, S., R.S. McAndrew, and K.W. Osteryoung. 2001. FtsZ ring formation at the chloroplast division site in plants. Journal of Cell Biology. 153(1):111-119.
Wächtershäuser, G. 2003. From pre-cells to Eukarya – a tale of two lipids. Mol. Microbiol. 47:13–22.
Wang, X., D. Ryu, R.H. Houtkooper and J. Auwerx. 2015. Antibiotic use and abuse: A threat to mitochondria and chloroplast with impact on research, health, and environment. Bioessays. 37(10):1045-1053.
Weijers, J.W.H., S. Schouten, E.C. Hopmans, J.A.J. Geenevasen, O.R.P. David, J.M. Coleman, R.D. Pancost, and J.S. Sinninghe Damsté. 2006. Membrane lipids of mesophilic anaerobic bacteria thriving in peats have typical archaeal traits. Environmental Microbiology. 8(4):648-657.
Wickstead, B. and K. Gull. 2011. The evolution of the cytoskeleton. J. Cell Biol. 194(4):513-525. www.jcb.org/cgi/doi/10.1083/jcb.201102065.
Williams, K.P., B.W. Sobral, and A.W. Dickerman. 2007. A Robust Species Tree for the Alphaproteobacteria. Journal of Bacteriology. 189(13):4578-4586.
Williams, T.A., P.G. Foster, T.M.W. Nye, C.J. Cox, and T.M. Embley. 2012. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc Royal Soc B. 279:4870-4879.
Woese, C.R. and G.E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 74 (11):5088–90.
Woese, C., O. Kandler, and M. Wheelis. 1990. "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya." Proc Natl Acad Sci USA. 87(12):4576–4579.
Yang, D., Y. Oyaizu, H. Oyaizu, G.J. Olsen, and C.R. Woese. 1985. Mitochondrial Origins. Proc. Natl. Acad. Sci. USA. 82:4443-4447.
Yutin, N. and E.V. Koonin. 2012. Archaeal origin of tubulin. Biol. Direct. 7:10.
Yutin, N., M.Y. Wolf, Y.I. Wolf and E.V. Koonin. 2009. The origins of phagocytosis and eukaryogenesis. Biology Direct. 4:9 doi:10.1186/1745-6150-4-9.
Zaremba-Niedzwiedzka, K., E.F. Caceres, J.H. Saw, D. Bäckström, L. Juzokaite, E. Vancaester, K.W. Seitz, K. Anantharaman, P. Starnawski, K.U. Kjeldsen, M.B. Stott, T. Nunoura, J.F. Banfield, A. Schramm, B.J. Baker, A. Spang, and T.J.G. Ettema. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 541:353-358.
Zubáčová, Z., L. Novák, J. Bublíková, V. Vacek, J. Fousek, J. Rídl, J. Tachezy, P. Doležal, C. Vlček, and V. Hampl. 2013. The mitochondrion-like organelle of Trimastix pyriformis contains the complete glycine cleavage system. PLoS One. 2013;8(3):e55417. doi: 10.1371/journal.pone.0055417.
How to cite:
Hegarty, S. 2018. Taxa: an overview of classification and major biological groupings. Mass Nature, https://massnature.com/taxa (25 November 2018).
